Statistics 410/510 Homework 3 Due: Friday, Feb.14
advertisement

1
Statistics 410/510
Homework 3
Due: Friday, Feb.14
Creating your own functions can be a useful feature in R when customizing your statistics. Functions are
especially useful when a procedure is repeated multiple times with different input. The basic structure
of a function is (1) a list of arguments that you provide the function and, (2) the output from the
function. The output can be graphs, numbers or both. Below are a few rudimentary examples of
functions in R that do addition, subtraction, and solve the quadratic function.
##############################
add <- function(a,b) # adds arguments a and b
{
cat("entering add function \n")
# \n is newline
cat("a=",a," and b=",b, "\n")
cat(a,"plus",b,"is equal to",a+b,"\n")
return(a+b)
# return()spits out value and ends the function
cat("function never reaches this line \n") # This is never touched
}
add(3,2)
#demonstrate
###############################
subtract <- function(a,b)
{
aminusb <- a-b # can create temporary values in functions
return(aminusb)
}
subtract(3,2)
aminusb
# objects don't exist outside of function
###############################
add.lazy <- function(a,b)
{
a*b # Ignored calculation, not seen later
a+b # Last calculation is spit out by function as though return() used
}
#############################
# A list can be used to return more than one item.
addsubtract <- function(a,b) # Demonstrate use of lists
{
aminusb <- a-b
aplusb <- a+b
# Using lists is useful when returning multiple values not in a vector
addminus <- list( amb=aminusb, apb=aplusb )
return( addminus )
}
addsubtract(3,2)
three.two <- addsubtract(3,2)
three.two$apb
# extracting an object from the list
#########################
2
quadraticFormula <- function(a, b, d )
# solve ax^2 + bx + d = 0
{
# Let d = c to avoid conflict with c() R command
# x1 = (-b-sqrt(b^2-4*a*c))/(2*a)
# x2 = (-b+sqrt(b^2-4*a*c))/(2*a)
if( (b^2-4*a*d) <0 )
{
cat("No real number solution - imaginary number solution needed \n")
return( NA )
}
else
{
x1 <- (-b-sqrt(b^2-4*a*d))/(2*a)
x2 <- (-b+sqrt(b^2-4*a*d))/(2*a)
return( unique(c(x1,x2)) )
# unique() returns only unique
# values - for when only one solution
}
}
# 2 solutions
quadraticFormula( 1, -2, -4 )
# solving
x^2 - 2x - 4 = 0
# No solution, imaginary numbers needed for solution, (b^2-4*a*c)<0
quadraticFormula( 3, 4, 2 )
# solving 3x^2 + 4x + 2 = 0
# 1 solution, sqrt(b^2-4*a*c)=0
quadraticFormula( 1, -6, 9 )
# solving
x^2 - 6x + 9 = 0
Lab Assignment
Create a function called mylm() which uses matrix algebra to perform a least-squares regression. The
results should match up with the output from R’s lm() function. (See example output to test your
results.) The function user will provide a vector for Y and a matrix for X. The provided X matrix will not
include the columns of 1’s for the intercept, so your function will need to create the column of 1s and
attach it to the front of the provided X matrix. Your function should return a list which includes:
a.
b.
c.
d.
e.
Estimates for the coefficients.
R-square
Adjusted R-square
Stat 510 students: Residual standard error
Stat 510 students: Standard errors for the coefficients
Begin the function by: mylm <- function(Y,X) {
your code here }
Remember that 𝛽̂ = (𝑋 𝑇 𝑋)−1 𝑋 𝑇 𝑦 and 𝑦̂ = 𝑋𝛽. Define the residual sum of squares (RSS) and total sum
of squares (TSS) as 𝑅𝑆𝑆 = ∑(𝑦𝑖 − 𝑦̂)2 and 𝑇𝑆𝑆 = ∑(𝑦𝑖 − 𝑦̅)2 . Also represent the number of variables
𝑅𝑆𝑆
2
(not counting the intercept) by p. Then 𝑅 2 = 1 − 𝑇𝑆𝑆 and the adjusted 𝑅 2 is 𝑅𝑎𝑑𝑗
=1−
𝑅𝑆𝑆⁄(𝑛−𝑝−1)
.
𝑇𝑆𝑆⁄(𝑛−1)
3
Roughly (but not always), an R-square closer to 1 is a good sign that the model describes the data well.
Because the R-square will only get larger (or stay constant) as more parameters are added (even if
nonsensible), the adjusted R-square penalizes the statistic a small amount for every parameter added to
the model. The residual standard error is 𝜎̂𝜀 = 𝑠𝜀 = √𝑅𝑆𝑆⁄(𝑛 − 𝑝 − 1) and
𝑆𝐸𝛽̂ = 𝜎̂𝜀 √𝑑𝑖𝑎𝑔[(𝑋 𝑇 𝑋)−1 ]
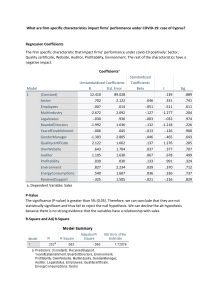
Example output:
> x <- c( 4,6,8 )
> y <- c( 2, 6, 9 )
> summary(lm(y~x))
Call:
lm(formula = y ~ x)
Residuals:
1
2
3
-0.1667 0.3333 -0.1667
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -4.8333
0.8975 -5.385
0.1169
x
1.7500
0.1443 12.124
0.0524 .
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.4082 on 1 degrees of freedom
Multiple R-squared: 0.9932,
Adjusted R-squared: 0.9865
F-statistic:
147 on 1 and 1 DF, p-value: 0.05239
> mylm(y,x)
$Beta
[,1]
-4.833333
X 1.750000
$RSquare
[1] 0.9932432
$RSquareAdj
[1] 0.9864865
$ResidualSE
[1] 0.4082483
$BetaSE
X
0.8975275 0.1443376
4
>
>
>
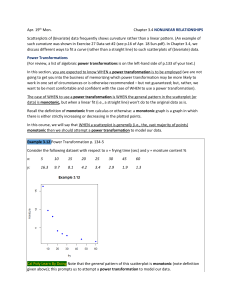
>
x <- cbind( c(4,6,8,10), c(5,3,6,7) )
y <- c( 2, 6, 9, 4 )
fit.lm <- lm(y~x)
summary( fit.lm )
Call:
lm(formula = y ~ x)
Residuals:
1
2
-0.8936 -0.4468
3
4
3.5745 -2.2340
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept)
4.0213
8.2629
0.487
0.712
x1
0.8617
1.3217
0.652
0.632
x2
-0.9149
1.9982 -0.458
0.727
Residual standard error: 4.332 on 1 degrees of freedom
Multiple R-squared: 0.2985,
Adjusted R-squared: -1.105
F-statistic: 0.2127 on 2 and 1 DF, p-value: 0.8376
> mylm(y,x)
$Beta
[,1]
[1,] 4.0212766
[2,] 0.8617021
[3,] -0.9148936
$RSquare
[1] 0.2984689
$RSquareAdj
[1] -1.104593
$ResidualSE
[1] 4.331969
$BetaSE
[1] 8.262938 1.321677 1.998188