Fig. S1 Fig. S1 Phylogenetic tree of S
advertisement

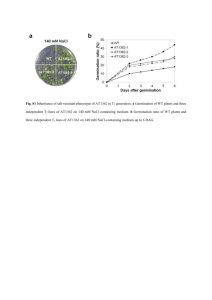
Fig. S1 Fig. S1 Phylogenetic tree of S-RBPs. The phylogenetic tree was made by ClastalW2 Phylogeny using default parameters. Fig. S2 Fig. S2 Salt stress response of S-RBP11 RNAi transgenic plants. (a) Schematic map of RNAi vector for the suppression of S-RBP11 and expression analysis of S-RBP11 in T1 RNAi lines. GAPc was used as an internal control. (b) and (c) Responses of WT and S-RBP11 RNAi T2 seedlings to 0, 130, or 140 mM NaCl. In (c), error bars represent standard deviation (n = 24 plants). Fig. S3 Fig. S3 Expression analysis of S-RBP11 under abiotic stresses using semi-quantitative RT-PCR. (a) Expression of S-RBP11 under 300 mM NaCl treatment for 0, 1, 2, 4, or 8 h. (b) Expression of S-RBP11 under cold treatment for 0, 1, 6, 12, or 24 h. (c) Expression of S-RBP11 under 10 μM treatment for 0, 1, 2, 4, or 8 h. In (a) to (c), GAPc was used as an internal control. Similar results were obtained from at least two biological replicates, with one shown here. Table S1 Primers used in this study Gene 35S enhancer copy 1 35S enhancer copy 2 35S enhancer copy 3 35S enhancer copy 4 Forward Reverse Purpose 5′-TATCTCGAGGATCCCCAACATGGTGGAGC-3′ 5′-CGCGTCGACTAGATATCACATCAATCCAC-3′ Cloning 5′-TATGTCGACGATCCCCAACATGGTGGAGC-3′ 5′-CGCAAGCTTTAGATATCACATCAATCCAC-3′ Cloning 5′-TATAAGCTTGATCCCCAACATGGTGGAGC-3′ 5′-CGCGAATTCTAGATATCACATCAATCCAC-3′ Coning 5′-TATGAATTCGATCCCCAACATGGTGGAGC-3′ 5′-CGCCTGCAGTAGATACACATCAATCCAC-3′ Coning LB1 5′-ATGGTCATAGCTGTTTCCTGTGTGAAATTG-3′ TAIL-PCR LB2 5′-AACCTGTCGTGCCAGCTGCATTAATGAATC-3′ TAIL-PCR AD1 5′-NGTCGASWGANAWGAA-3′ TAIL-PCR AD2 5′-TGWGNAGSANCASAGA-3′ TAIL-PCR AD3 5′-AGWGNAGWANCAWAGG-3′ TAIL-PCR AD4 5′-WGTGNAGWANCANAGA-3′ TAIL-PCR LB-seq 5′-CATTAATGAATCGGCCAACG-3′ Sequencing S-RBP11 5′-ATCGTTTTGCACTACAGAGC-3′ 5′-CACGAAACCGAAACCTTTTG-3′ RD29A 5′-CCTGAAGTGATCGATGCACC-3′ 5′-CAGTGGGTTTGGTGTAATCG-3′ COR15A 5′-AACGCAAGAAGTCGTTGATC-3′ 5′-CGTTGAGGTCATCGAGGATG-3′ GAPc 5'-GTGTCCCAACCGTTGATGTC-3' 5'-TCCCTTGAGTTTGCCTTCGG-3' S-RBP11 At5g06190 5'-AAATTTTAATGGCGGCGCTC-3' 5′-TACAGCTCGCGATCCAGATC-3′ 5'-TGCGTAGTCCACAAATATCG-3' 5′-ATTATTCCTCTAGCCCTCAG-3′ Quantitative RT-PCR Quantitative RT-PCR Quantitative RT-PCR Quantitative RT-PCR Semi-quantitative RT-PCR Semi-quantitative RT-PCR Semi-quantitative GAPc 5'-CACTTGAAGGGTGGTGCCAAG-3' 5'-CCTGTTGTCGCCAACGAAGTC-3' Entire S-RBP11 5'-ATATCTAGAATGGCGGCGCTCGCA AGGAT-3' 5'-TATCCCGGGGTCGCTTTTCGATGTTTCGG-3' Cloning 5'-CCGTCTAGAATGTCCGCCTCATGTTTCTTC-3' 5'-TATCCCGGGGTCGCTTTTCGATGTTTCGG-3' Cloning 5'-ATCCTGCAGATGGTGAGCAAGGGCGAGGAG-3' 5'-ATCGTCGACCTTGTACAGCTCGTCCATGCC-3' Cloning 5′-GAAGATCCAGCGAAAAGATGGAC 5′-GATCCAAAAAAACGGACAGCAATTAA GGCTGTTCAAGAGACAGCCGTCCA ACGGCCGGACGTCTCTTGAACGTCCGG TCTTTTCGCTGGATCTTTTTTTG-3′ CCGTTTAATTGCTGTCCGTTCTGCA-3′ Truncated S-RBP11 sGFP S-RBP11 RNAi RT-PCR Cloning Table S2 List of small RNA-binding proteins in Arabidopsis S-RBP Protein code AGI code aa length Domain S-RBP1 CAB78428, NP_193122 At4g13860 126 One RRM S-RBP2 BAB09544, NP_200183 At5g53720 100 One RRM S-RBP3 AAC61284, NP_179093 At2g14870 101 One RRM S-RBP4 CAB88326, NP_190188 At3g46020 102 One RRM S-RBP5 CAB78306, NP_193001 At4g12640 823 Two RRMs and SPOC domain S-RBP6 AAC36547 S-RBP7 AAG51218 , NP_973958 At1g33470 245 One RRM S-RBP8 AAD20390, NP_179762 At2g21690 117 One RRM S-RBP9 AAC23648, NP_181287 At2g37510 142 One RRM S-RBP10 AAF21210, NP_187457 At3g08000 143 One RRM S-RBP11 BAB09686, NP_196239 At5g06210 146 One RRM S-RBP12 BAB09337, NP_200269 At5g54580 156 One RRM S-RBP13 BAB08354, NP_200794 At5g59860 157 One RRM S-RBP14 AAB88654, NP_850366 At2g42240 292 Two RRMs S-RBP15 BAB09540, NP_200179 At5g53680 169 One RRM alcohol dehydrogenase, partial (Fragaria viridis)