File - Middle School Science Lesson Plans
advertisement

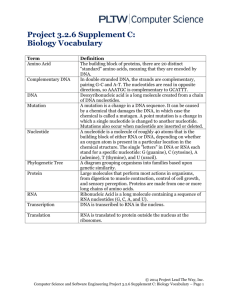
Lesson Plan Template Name of Lesson/Unit: DNA Barcoding Grade Level: 10-12 Prepared by: Section 1: Lesson Summary Topic: Evidence of molecular heredity and evolution NGSS Performance Expectation: HS-LS3-2: Make and defend a claim based on evidence that inheritable genetic variation may result from 1) new genetic combinations from meiosis, 2) errors in replication, or 3) mutations. Crosscutting Concept: Cause and Effect – Empirical evidence is required to differentiate between cause and correlation and make claims about specific causes and effects. Engineering Practices: Analyzing and interpreting Data – formulating, refining, and evaluating empirically testable questions and design problems using models and simulations. Common Core State Standards (Reading): “Barcode of Life” by M. Stoeckle and P. Hebert, Scientific American, October, 2008, pp. 82-88. RST.11-12.1 Cite specific textual evidence to support analysis of science and technical texts, attending to important distinctions the author makes and to any gaps or inconsistencies in the account. Common Core State Standards (Writing): WHST.9-12.1 Write arguments focused on discipline-specific content. Students will prepare a written defense of research communicating the analysis and conclusions of their research. Current CA Science Standards: Students know how mutations in the DNA sequence of a gene may or may not affect the expression of the gene or the sequence of amino acids in an encoded protein. Students know new mutations are constantly being generated in a gene pool. Objective of the Lesson or Unit: Students will observe, record and analyze molecular evolution and predict semiquantitatively how this may affect phenotype (e.g., DNA nucleotide differences and amino acid changes in the gene product). How is this objective relevant to students? Students will be completing a research project and writing a research paper for publication. Students will be participating in a worldwide scientific initiative to barcode 5 million specimens representing 500,000 different species and make the data available to the scientific community in the International Barcode of Life (iBOL) database. Culminating Assessment (How will you know students have met the Performance Expectation?) Describe the assessment and attach a copy. Students will produce a research paper for publication. Students will present a defense of their research to the Science Magnet faculty and invited scientists. Lesson Plan Template What has been taught in my class to prepare students for this topic? DNA replication, transcription, translation, protein structure and function, cellular respiration, mechanisms of evolution, natural selection, experimental approaches and techniques used in molecular life science Description of the Lesson/Unit (Use the following Table) With 5 E and How Short description of activity People Learn in Mind! Engage Students join the iBOL DNA barcoding scientific initiative Explore Students collect specimens, extract total DNA, amplify COI gene, purify amplicon by spin column chromatography, use automated DNA sequencing to determine 650 bp vertebrate COI barcode amplicon Explain Students will analyze DNA barcodes of specimens: create phylogenetic tree, identify intraspecific and interspecific nucleotide differences of specimen barcodes; identify codon changes, determine nucleotide position within the codon, determine amino acid Purpose/Intention for the activity or Key Learning How does this activity build student knowledge and skills towards meeting the Performance Expectation? Identify at least 1 Practice Students discover the value of species identification and taxonomy in order to preserve and utilize the biodiversity of the biosphere and how they can contribute to the scientific community through DNA barcoding; students develop an understanding of how molecular evolution contributes to speciation Students carry out molecular life science protocols to determine the DNA barcodes of their specimens; students are motivated to understand the concepts of DNA replication, transcription, translation, oxidative phosphorylation and the techniques of molecular life science (nucleic acid isolation and purification, polymerase chain reaction PCR and DNA sequencing) which are relevant to their research Students gain an in depth understanding of the expression of genetic material, the nature of evolutionary mutations and their effects on gene products, the molecular basis for speciation as depicted in a phylogenetic tree, and the power of computational biology in analyzing large amounts of data; students create Lesson Plan Template Elaborate Evaluate produced in the gene product, quantify these changes, and determine their effect on the gene product Students create a scientific research paper for publication in which they communicate the findings of their research Students will present a defense of their research to the Science magnet faculty and invited scientists specimen pages and sequence pages from their research on the eBOL database which appears on NCBI after teacher and scientist vetting. Students analyze and incorporate primary resources in communicating their analyses and conclusions of their research Defending their research in a high stakes situation will allow both the students and the teacher to assess the extent to which students have developed in depth understanding of content, that students reached logical and evidence based conclusions, and the effectiveness of communicating their findings with the academic and scientific community Section 2: Laboratory/ Hands-on Lesson Content Objective: Students will use bioinformatics to identify mutations in DNA barcodes of rockfish species as evidence of evolution, and will analyze the effect of mutations on the gene product; students will construct a phylogenetic tree illustrating the evolutionary relationships among the DNA barcoding specimens Language Objective: Students will become fluent in the use of relevant biological terms related to DNA barcoding and their research. Assessment: How will I know students met the objective? Students will prepare a scientific manuscript for publication which will be submitted to an appropriate journal (National High School Journal of Science, Journal of Experimental Secondary Science, Journal of Youths in Science, Torrey Pines Falconium Journal, etc.) Rubric: Assessment criteria 1. Students follow the format for the journal to which they submit their manuscript. 2. Students introduce the research topic and provide sufficient background information for general understanding. 3. Students clearly state the questions and/or hypotheses being addressed in the research. 4. Students describe the experimental procedures utilized to address the questions/hypotheses. 5. Students present the data in appropriate tables. 6. Students use appropriate analytical and statistical methods to interpret data. 7. Students thoroughly discuss and explain research results, relating them to current scientific theories and applications. Lesson Plan Template Description of lesson: Bioinformatics of DNA barcodes to identify mutations and analyze their effects on the gene product, COI. Students use BLAST to align DNA barcodes of class specimens to identify nucleotide changes; students quantify the codon positions in which they occur and the type of base substitution (A→G, C→T, etc.); students determine the types of amino acid changes that occur due to nucleotide changes; students identify the locations of amino acid changes in the protein sequence of COI Time required for the lesson: 2 hour introduction of bioinformatics tools (BLAST, nucleotide changes, open reading frame (ORF), vertebrate mitochondrial genetic code for sequence translation; 2 hours of student analysis; 2 hours of homework to communicate findings for manuscript Materials/ Preparation needed: DNA barcode sequences on BOLD-SDP (www.boldsystems.org), computers with Excel Description of Student Task (each step) Time Needed for Students to complete each Task Use BLAST to align class DNA barcodes; use aligned sequences to identify and tabulate nucleotide differences 1 hr. class, 1 First 15 minutes: have students hr. attempt to manually align DNA homework barcode sequences Introduce power of computational biology to align sequences instantaneously (15 min) Have students select DNA barcodes from BOLD-SDP and align (15 min) Have students brainstorm efficient strategies to tabulate nucleotide changes (15 min): check for understanding. 1 hr. class, 1 These concepts have been hr. covered previously in a research homework project to clone and express a Quantify the different types of base substitutions and the Teacher Notes (Include strategies/questions to check for understanding) Accommodation/Mo dification for Student Access and Extension (i.e. strategies for EL and extensions for GATE) EL: check for understanding of relevant vocabulary GATE: addition of statistical analysis of data to determine pvalues EL: check for understanding of relevant vocabulary Lesson Plan Template codon positions in which they occur; identify the open reading frame (ORF) in order to identify the amino acid changes produced by the nucleotide changes Analyze the frequency of changes in different codon locations and open up a discussion to interpret the findings; analyze the types of nucleotide changes and whether they produce a change in the amino acid of the gene product; open up a discussion of the types of amino acid changes which occur and where in the gene product they occur gene and then isolate the gene product; review, relate and check for understanding 1 hr. class time for nucleotides, 1 hr. class time for amino acids Nucleotide changes: What types of changes occur most often (usually purine for purine or pyrimidine for pyrimidine); In which position of the codon do most nucleotide changes occur (3rd, then 1st, and rarely 2nd) – initiate class discussion (students usually know and apply wobble hypothesis); What are the different classes of amino acids (polar, nonpolar, acidic, basic); How do amino acids affect the structure and function of a protein (polarpolar, nonpolar-nonpolar, and acidic-basic interactions affect protein folding/conformation which determines function) GATE: investigate the different genetic codes; compare and contrast EL: check for understanding of relevant vocabulary GATE: using PDB, identify regions of COI in which nucleotide changes occur/don’t occur Section 3: Science Literacy Content Objective: Using nucleotide changes in specimen DNA barcodes (Channel Island rockfish), apply parsimony and nearest neighbor analysis to create a phylogenetic tree for the class species Language Objective: Students will become fluent in the use of relevant biological terms related to DNA barcoding and their research. Lesson Plan Template Assessment: How will I know students met the objective? Students will prepare a scientific manuscript for publication which will be submitted to an appropriate journal (National High School Journal of Science, Journal of Experimental Secondary Science, Journal of Youths in Science, Torrey Pines Falconium Journal, etc.) Rubric: Assessment criteria 1. Students follow the format for the journal to which they submit their manuscript. 2. Students introduce the research topic and provide sufficient background information for general understanding. 3. Students clearly state the questions and/or hypotheses being addressed in the research. 4. Students describe the experimental procedures utilized to address the questions/hypotheses. 5. Students present the data in appropriate tables. 6. Students use appropriate analytical and statistical methods to interpret data. 7. Students thoroughly discuss and explain research results, relating them to current scientific theories and applications. Description of lesson: Students will use the nucleotide differences identified in the specimen DNA barcodes to construct a phylogenetic tree, and will analyze the evolutionary relationship of the included species and instances of speciation in progress Time required for the lesson: Materials/ Preparation needed: DNA barcode sequences on BOLD-SDP (www.boldsystems.org) Description of Student Task (each step) Students use tabulation of nucleotide differences to quantify intraspecific and interspecific differences; students create phylogenetic trees (rooted and unrooted) and Time Needed for Students to complete each Task 1 hr. class Teacher Notes (Include strategies/questions to check for understanding) Accommodation/Modification for Student Access and Extension (i.e. strategies for EL and extensions for GATE) Allow students to begin to grind through the analysis (30 min) and then demonstrate the power of computational biology to produce the phylogenetic tree instantaneously; What relationships do we see EL: check for understanding of relevant vocabulary GATE: write computer programming code to rank number of nucleotide differences between specimens Lesson Plan Template phylograms using bioinformatics Analyze the phylograms to determine evolutionary relationships and instances of speciation in progress 1 hr. class Compare the number of nucleotide differences within a species to the number between different species (fewer differences within a species <2%, more differences between species >2%); How can the process of speciation be identified (on a phylogram, specimens within a species begin to approach the 2% difference); What processes can lead to speciation (geographic isolation, reproductive isolation, etc.) Section 4: Student Pages Please provide a printer/copier friendly student pages for Section 2 and Section 3.