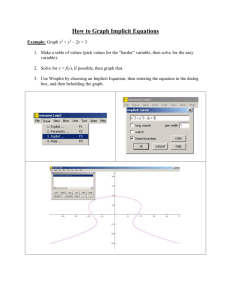
Cooper,Herskovits
advertisement

NOTES on Cooper,Herskovits 1992 Machine Learning: Bayesian network scoring
Notation
ci Î{v i1,...,v ir }, possible values of node c i
i
qi = # configurations (w ij ) of p i , the parents of ci
D = the data = {c ih }; h indexes cases
BP ijk = Pr(ci = v ik | p i = w ij ) = the model
Nijk = #{h : c i = v ik & p i = w ij )
To make it simple, consider just a single variable i and a single parent configuration j. The
number of values is ri º K , data are (Nij1,...,Nijr ) º (n1,...,nK ) , multinomial with sample size
i
Nij º n and parameter vector (BPij1,...,BPijr ) º (p1,...,pK ) . The marginal probability of the
i
data is dirichlet-multinomial. When K = ri =2, the marginal of n1 is beta-binomial, and
with a flat prior it is discrete uniform on {0,…,n} and of course n2 = n - n1 . Generally,
Pr(n1,...,nK ) =
=
n!
ò
p
ò Õn !Õ p
nk
k
Pr(n1,...,nK | p1,...,pK )p (p1,...,pK )
×1/ Dir(1,...,1)
k
(1)
=
=
n!
n
Dir(n1 + 1,...,nK + 1) × pk k / Dir(1,...,1)
Õ nk !
n! Õ G(nk + 1)
×(K - 1)!
Õ nk ! G(K + n)
n!
×(K - 1)!
(K + n - 1)!
This is the reciprocal of the number of distinct values for the multinomial. Thus the
probability is uniform across all data vectors. This we confirm by simulation. For K =2,
this is 1/(n+1), a well known result for beta-binomial.
=
This is for a specific ordered data vector, where you know the count for each label v ik .
The C-H formula is:
Õ
nk !
×(K - 1)!
(K + n - 1)!
which you obtain by omitting the multinomial coefficient.
(2)
If you know only the set of counts but not the order (which count goes with which
variable), the number of sets is given by the C-H formula. If the different data vectors
leading to the same count vector are counted as 1 instead of n!/
nk !
,
indeed you would divide by n!/
nk ! and get agreement with C-H.
Õ
Õ
This is similar to Bose-Einstein statistics, in which the indistinguishability of bosons
means that, for example, three photons distributed to two energy levels have probability
distribution
Pr(HL)=Pr(HH)=Pr(LL)=1/3
instead of
Pr(HL)=1/2, Pr(HH)=Pr(LL)=1/4.
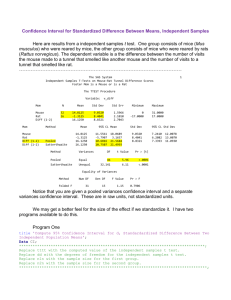
Demonstration that (1) is correct by simulation is seen in Cooper,Herskovits.html.
Going from A4 to A5:
To simplify, n=1, and qi=1, and ri=2.
A4 amounts to
m
Õp
dh
1-dh
(1- p)
= pn (1- p)m-n , corresponding to a single data assignment.
h=1
A5 is the same. Near the bottom of p.33, C-H distinguish between D and D’, ordered and
unordered data results. So I think everything’s fine, since they deal with D and I am dealing
with D’.
Next issue:
For each parent configuration w, the probability of the data, conditioning on the parent
but marginalizing on the probability table, will be uniform on count vectors D’ as we have
shown. In terms of D, the probability will be lower for count vectors with lots of
multiplicity, i.e. where n!/
nk ! is big. For example, for 8 Bernoulli observations, the
Õ
probability is highest for all heads or all tails, and smallest for 4 each regardless of order.
The process penalizes impure nodes. Very close to classification trees it seems.
(Note: The uniform dirichlet density is 1/(K-1)! = 1/D(1,…,1).
The volume of the simplex is NOT 1/(K-1)! For example, for K=2, the 1-simplex has length
sqrt(2), but the density is 1, not 1/sqrt(2). This is because of the Jacobean, transforming
between (p1,p2 ) and (p1 - p2 ,p1 + p2 ) , yielding sqrt(2) ; K=3, the 2-simplex has base =
sqrt(2) and height = sqrt(sqrt(2)^2-(sqrt(2)/2)^2)=sqrt(3)/2. —Jacobean!)