tpj12798-sup-0010
advertisement

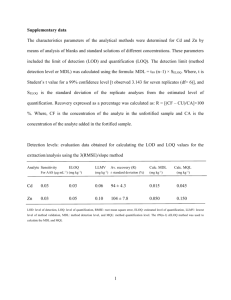
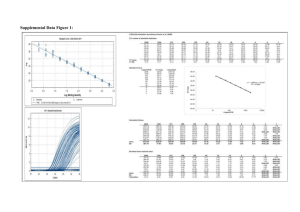
Data S1: Limits of detection (LOD) and quantification (LOQ) of ME-glucosides and ion suppression of MEP pathway intermediates ME-glucosides Retention time, extraction efficiency, limit of detection (LOD), limit of quantification (LOQ) and the absolute amounts of MEGlu in wild type Arabidopsis thaliana leaf extracts. The extraction efficiency values represents the average ± SE (n = 3). Due to the unavailability of isotopically labelled standards for ME-glu and the absence of 13C label incorporation during short term 13CO2 labelling, the extraction efficiency and absolute amounts were determined using a standard addition curve. For the standard addition curve a similar amount of both isomers were added in triplicate at three known amounts to approximate 75%, 150% and 200% of the ME-glu amount as calculated using an external standard curve. The LOD was taken as the amount of metabolite detected that gave a signal to noise (S/N) ratio of 3 and the LOQ was taken as the amount necessary to give an S/N ratio of 5. Retention time (min) Extraction efficiency (%) LOD (pmol ml-1) LOQ (pmol ml-1) Absolute amounts (nmol mg-1 DW) MEGlu 4.0 14.8 ± 0.75 44.9 ± 1.9 74.8 ± 3.2 2.639 ± 0.09 MEP pathway intermediates Retention time, extraction efficiency, ion suppression, limit of detection (LOD) and limit of quantification (LOQ) of DXP, MEcDP and DMADP/IDP in Arabidopsis thaliana leaf extracts. The extraction efficiency and ion suppression values represents the average ± SE (n = 10). The LOD was taken as the amount of metabolite detected that gave a signal to noise (S/N) ratio of 3 and the LOQ was taken as the amount necessary to give an S/N ratio of 5. Retention time (min) Extraction efficiency (%) Ion suppression (%) LOD (pmol ml-1) LOQ (pmol ml-1) DXP 6.9 54.0 ± 2.6 105.5 ± 5.7 7.8 ± 0.2 13.0 ± 0.3 MEcDP 6.0 47.8 ± 2.5 55.5 ± 4.0 7.0 ± 0.03 11.6 ± 0.04 DMADP/IDP 6.6 14.2 ± 1.0 76.5 ± 1.3 1.4 ± 0.01 2.3 ± 0.01