Shaun Lynn Protein report
advertisement

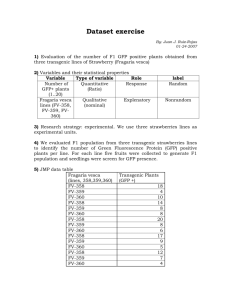
RECOMBINANT DNA TECHNIQUES: GFP ANALYSIS USING CHROMATOGRAPHY, INDUCING GFP IN E.COLI CELLS AND SDS PAGE ANALYSIS. SHAUN LYNN APPLIED CHEMISTRY – J9106574 Abstract A cDNA fragment encompassing the Green Fluorescent Protein encoding gene (GFP) was introduced to an E.Coli sample. Positive and negative cultures were developed and tested for UV fluorescence. The positive sample was then subjected to hydrophobic interaction chromatography, with the eluted sample being collected in three tubes and then tested for fluorescence under a UV light. The transformed E.Coli was then prepared for inducing the expression of GFP and the optical densities were recorded using a synergy HT micro plate reader. The induced GFP was then extracted and analysed using SDS PAGE analysis Introduction What is recombinant DNA Recombinant DNA is defined as “a fragment of DNA incorporated artificially into the DNA molecule of another organism to enable multiple replication of itself. This way a large quantity of the DNA in question can be obtained (Turner, 2006) Recombinant DNA can be formed using three routes. Transformation; using bacterial cells such as E.coli. Phage introduction, very similar to transformation except a phage is used as an alternative host, and finally Non-bacterial Transformation, which is similar to transformation, the only real difference is that it does not use bacteria. Instead the DNA is injected directly into the nucleus of the cell that is to be transformed. (An Introduction to Recombinant DNA, 2011) Applications Recombinant DNA technology is vital in modern scientific research, it has also altered the way we diagnose and treat genetic disorders and diseases. Identification of mutations. People may be tested for the presence of mutated proteins that may be associated with cancer or illnesses such as neurofibromatosis. . Tests exist to determine if people are carriers of the cystic fibrosis gene and the Huntington's disease gene. Genes can be transferred from one organism to another. Sufferers of cystic fibrosis, rheumatoid arthritis and certain cancers find this kind of gene therapy extremely beneficial By mapping human genes on chromosomes, mutations can now be pin pointed to specific sites on chromosomes. Research conducted by the University of Georgia USA states that the other benefits of Recombinant DNA technology include the production of insulin, cloning and chimeric plasmids”. (Unknown, 2011) Advantages and drawbacks of recombinant DNA technology Recombinant DNA technology is a fast and efficient way to amplify specific DNA sequences from minute amounts of sample material, without the need to clone or sub-clone. Disease diagnosis “will be greatly expedited by PCR to identify microorganisms in infected people who would prove falsely negative by other diagnostic procedures.” (Unknown, BIO 102 Biological Sciences, 2011) During the amplification process, this technology can introduce errors into the DNA in addition to this, DNA polymerases have been found to be error prone. Although this is quite infrequent, new enzymes such as Vent polymerase and Taq polymerase dramatically reduce this problem. Another major factor to consider is the introduction of contaminants, minute quantities can yield false positives or erroneous results. Chromatography Chromatography is the name given to the analytical technique that involves the separation of mixtures. It involves passing a solution, dissolved in a specific mobile phase through a specific stationary phase. This separates the analyte, allowing it to be measured against different molecules in the solution, based on the differential separation of the mobile and stationary phases. The degree of adsorption is dependent on polarity, non polar compounds, such as alkanes, Olefins are weakly adsorbed and will therefore be easily eluted. Polar compounds, such as alkyl halides, alcohols and amines however are strongly adsorbed and will be very difficult to elute. Chromatography may therefore be used as a form of purification, by separating the required components of a mixture from the unwanted for further use. Column chromatography is the most frequently used separation technique in which the stationary bed is within a tube. (McMurry, 2008) Electrophoresis Electrophoresis is a method used for the separation of molecules in mixtures by use of applied electric field. This specific electric field separates molecules according to mobility. The molecules dissolved within the mixture move across ions according to their charge and mass ratio e.g. a molecule with the bigger net charge will move faster to an electrode. Page electrophoresis separates molecules according to their molecular mass which is directly proportional to the voltage and charge of the molecule. . (Weaver, R. F., (2008).) (Unknown, 2011) Aims To analyse expression of GFP using chromatography. To measure the band size of the GFP Protein in SDS Page analysis. To find the average optical density of E. coli after it was induced with GFP by plotting Absorbance over time. Method- GFP Analysis using Chromatography Using a UV lamp, several isolated green colonies on the LB/amp/Ara plate were identified. Several isolated white colonies on the LB/amp plate were then isolated. One of the LB/Amp/Ara growth media tubes were labelled positive and the other labelled negative. A sterile loop was used to transfer one green colony to the positive growth medium tube. The loop was spun to disperse the colony. A new sterile loop was used to transfer one white colony to the negative growth medium tube and was once again spun to disperse the colony amongst the medium. The two tubes were placed in a rotating incubator (37°C) for one hour. After the set time the liquid cultures were removed from the incubators and observed under a UV lamp, any differences in colour were noted. The negative growth medium was then discarded. The positive growth media (2ml) was transferred to a microcentrifuge tube and labelled ‘positive’. It was then placed in a centrifuge for 5 minutes at 14,000xg; the supernatant was then removed without disturbing the pellet. The pellet was then observed under UV light, any observations were noted. TE buffer (250μl) was added to the tube and the pellet was resuspended by gentle pipetting. Lysozome (250μl) was added to the sample and mixed gently. The tube was observed under UV light and any observations were noted. The samples were incubated on ice for 15 minutes. They were then centrifuged for 10 minutes at 14,000xg. The cap was removed from the chromatography column and the bottom was snapped, allowing the liquid contained inside to drain. Equilibrium buffer (2ml) was added to the column and allowed to drain down to the 1ml marker; the column was then capped at the top and bottom. The sample was then removed from the centrifuge and observed under UV light, any observations were noted. 250μl of the supernatant was transferred into a clean microcentrifuge tube, binding buffer (250μl) was added and the sample was then placed on ice until needed. Three collection tubes were labelled 1-3 and placed in the foam rack. The cap was removed from the chromatography column and placed into collection tube 1; the liquid was allowed to drain until it was level with the top of the matrix. The column was then carefully loaded with 250μl of the sample. The column was examination under a UV lamp. When all the liquid had drained, the column was then transferred to collection tube 2. Wash buffer (250μl) was added to the column and the entire volume was allowed to drain. The column was examined again under UV light, noting any observations. The column was then transferred to collection tube 3. TE buffer (750μ) was added to the column and the entire volume was allowed to drain through the matrix. All three tubes were observed under UV light, observations were noted. They were then parafilmed and placed on ice. Method- Inducing Expression of GFP in E.coli *overnight cultures of transformed E.coli at mid-stationary phase were provided* The overnight culture (5ml) was added into a pre warmed LB medium to create a 1:10 subinoculum. Two 1ml samples of the sub-inoculated culture were pipetted into two separate 1.5ml Eppendorf tubes. They were labelled with the group number and sample time e.g.T0 and then placed on ice. The sub-inoculated culture was incubated at 37°C with a shaking rate of 200rpm. This process was repeated every 30 minutes for each sample, ensuring the samples were placed on ice afterwards. After 60minutes, arabinose (1.5ml) was added. The sub-inoculated culture and arabinose were incubated at 37ºC with shaking rate of 200rpm. Every 30 minutes, the sub-inoculated culture was incubated at 37°C with a shaking rate of 200rpm with the additional arabinose. This was repeated until the sampling time reached T180. From this, two samples for each sampling time had been procured; one set was placed on ice and set aside for protein analysis in a later experiment. For the second set of samples, 200μl of each sampling time was taken and loaded into three separate wells of a UV flat bottomed transparent 96 well plate according to the plate layout provided. 200μ of LB broth was used for the blank wells. The plate was placed into the Synergy-HT Micro plate reader and measured for the optical density of the samples at 600nm. The optical density values were recorded, an average was calculated for each sampling time and a graph of absorbance vs. time was plotted. Extraction of induced GFP from E. coli and SDS-PAGE Analysis of Extracted GFP The samples were centrifuged for 5 minutes at 14,000xg in order to harvest the bacterial cells. The supernatant was discarded ensuring the pellet was not disturbed. Protein extraction buffer (50μl) was added and the pellet was resuspended using a vortexer. The samples were incubated in boiling water for 5 minutes. The samples were then centrifuged form 5 minutes at 14,000xg to clarify the samples. The supernatant was carefully removed and placed into a clean 1.5ml Eppendorf tube and placed on ice to cool. SDS-PAGE Analysis of Extracted GFP 20μl of molecular weight ladder was loaded to the first well of the gel. The protein samples were then loaded into the well according to the gel layout. The gels were ran at 200V, until the dye front was approximately 1cm from the gel bottom, this took roughly 1hour. The gel cassette was carefully disassembled and placed into a staining tray. The gel was washed with distilled water (100ml). The water was removed and Comassie Blue (0.5 %( w/v)) (100ml) was added. The gel was then left for 1 hour to stain on a rotating bed. The stain was removed and rinsed thoroughly with distilled water. Destain solution (100ml) was then added and then left overnight. The destain solution was then replaced with fresh destain solution (100ml). The gel was further destained until the protein bands were visible. The destain was then removed and rinsed thoroughly with distilled water. The gel was then visualised using the Alpha Innotech Gel Doc System. Results GFP analysis observation table Step number 7 11 13 18 25 28 31 32a 32b 32c Item exposed to UV light +ve Culture -ve Culture Pellet Cell suspension +Lysozyme Centrifuged samples Column with sample Column with sample and wash buffer Column with sample and TE buffer Collection tube 1Sample in binding buffer Collection tube 2Sample in Wash buffer Collection tube3Sample with elution buffer Prediction Will fluoresce Will not fluoresce Will Fluoresce Will not fluoresce Observation under UV light Fluoresced green Did not fluoresce Fluoresced green Did not fluoresce Will not fluoresce Did not fluoresce Will Fluoresce Fluoresced green Will not fluoresce Did not fluoresce Will not fluoresce Did not fluoresce Will not fluoresce Did not fluoresce Will not fluoresce Did not fluoresce Will Fluoresce Fluoresced green The results of Chromatography at 10 μl showing the expression of the GFP in collection tubes 1, 2 and 3 Figure 1- The GFO fluoresced the most in collection tube three, Where GFP expression was at its highest. The results of Chromatography at 20 μl showing the expression of the GFP in collection tubes 1, 2 and 3 FIGURE 2 - THE GFP FLOURESCED THE MOST IN COLLECTION TUBE THREE, WHERE GFP WAS AT ITS HIGHEST Averages of Absorbance Readings Inducing GFP in E. Coli-single readings for each sample Average T0 T30 T60 T90 T120 T150 T180 600 0.17567 0.16333 0.19833 0.252 0.323 0.417 0.513 620 0.167 0.156 0.18833 0.238 0.26633 0.33433 0.477 640 0.16 0.14933 0.17467 0.21567 0.296 0.369 0.444 660 0.154 0.14367 0.17467 0.21567 0.28333 0.36233 0.442 680 0.14867 0.13767 0.16967 0.21 0.27567 0.35833 0.441 700 0.14433 0.13267 0.16433 0.20533 0.26833 0.35 0.43367 Total average 0.158 0.147 0.178 0.223 0.285 0.365 0.458 Time 0 30 60 90 120 150 180 TABLE 1 AVERAGE ABSORBANCE READINGS Average of singular absorbance readings against time of inducing GFP in E.coli cells Average Optical Density (Absorbance Units) 0.5 0.45 0.4 0.35 0.3 0.25 0.2 0.15 0.1 0.05 0 0 20 40 60 80 100 Time (s) 2-1 THE HIGHLIGHTED POINT INDICATES THE ADDITION OF ARABINOSE 120 140 160 180 200 The expression of GFP through the method of SDS PAGE gel electrophoresis The SDS PAGE Electrophoresis shows arabinose was mostly present at 60KDA, 50KDA and 40KDA suggesting that the average KDA for GFP expression was 50KDA. This is where GFP expression was at its highest. It was discovered from the SDS PAGE electrophoresis that the band size in which the GFP expression was at its highest was at 50 KDA. This was measured by taking an average of three bands where GFP expression was at its highest and discovering the mean expression. Discussion From the observations of the chromatography practical, it was clear to see that only collection tube three fluoresced, this will be due to the fact that it was only in this tube that GFP was expressed. The reason for this could be that the GFP structure contains many hydrophobic amino acids, which makes the entire GFP protein hydrophobic, overall the GFP protein will be more hydrophobic than the proteins present in E.coli. The beads in the column are extremely hydrophobic, when the bacterial lysate was added to the column; the hydrophobic proteins in the sample will interact with the stationary phase beads, whilst the hydrophilic proteins will be eluted. The presence of a buffer with a high salt concentration strengthens this interaction. . When the low salt buffer was added to the column, the hydrophobic core was no longer exposed and the proteins hydrophilic shell would allow the protein to be eluted in a purified more concentrated form. As UV Fluorescence is not the normal phenotype for E.coli. The PGlo plasmid DNA was not responsible for the bacterial UV fluorescence so therefore it is the Result of the new protein the E.coli. are producing the GFP protein. The results of the GFP induction suggest that the introduction of arabinose at time T60 increased the values of the average absorbance readings. This will be due the arabinose Inducer interacting with the E.coli codons, which will encourage the E.coli to express the GFP. The graph indicates there is a direct positive correlation between the increase in arabinose and the intensity of GFP expression. The role of the binding buffer in this practical was to control and regulate the pH of the sample. The role of the Wash buffer was to cleanse and lubricate the second collection tube whilst also removing debris from the chromatography column. The role of the TE elution buffer was to stabilize the E.coli DNA whilst protecting the DNA from denaturisation at the same time. (Watson, J., 2008) The function of the equilibration solution was to alter the salinity content to induce equilibrium in the column, this is an efficient way to regulate the amount of fluid being eluted from the. The function of the lysozyme was to destroy the bacterial cell wall initiating enzymatic digestion. The function of centrifuging was to break the cell membrane, by applying a detergent to create a sample called a homogenate. This mixture is then added to two tubes and attached to a rotor which is spun at high speed. In doing so, the cell components separate, more dense components sink and sometimes form a pellet, whilst the less dense components will rise to the top. The hydrophobic interaction matrix is where some proteins are hydrophobic and will repel water. The high salt concentration causes the GFP to fold over and expose the hydrophobic regions to the outside so the protein sticks to the column. (Wolfe, S. L., 1993). SDS page analysis is a detergent which causes proteins to denature without breaking their peptide bonds. SDS is added to the denatured protein sample to make it negatively charged. The hydrocarbon tails of the SDS can then bind to hydrophobic areas of the protein sample and the electric field between the SDS molecules which are already bound, forces the proteins to unfold. The SDS then molecules bind to the polypeptides. The speed at which the polypeptides travel down the gel is largely dependent on the pore size of the gel matrix and the weight of the protein. As larger polypeptides travel to the bottom of the matrix first, it is easier to measure individual weight and size of polypeptides in this particular way. (Schleif, R. F. Wensink, P. C, 1981) Arabinose is introduced to make the GFP function. There is a positive correlation between the addition of arabinose and the production of GFP; this will produce a larger band width and higher optical density. The intensity of the fluorescing GFP indicates a higher concentration of GFP, Suggesting the GFP expression rate can be measured by monitoring the intensity of the fluorescence against time. In addition to this, gel electrophoresis is a method that could be employed to measure GFP, as it separates proteins in accordance to their mobility, whilst SDS PAGE electrophoresis separates proteins according to their size. WORKS CITED An Introduction to Recombinant DNA. (2011, march 13). Retrieved http://rpi.edu/dept/chem-eng/Biotech-Environ/Projects00/rdna/rdna.html March 13, 2011, from Clark, J. (2011, 03 14). chromatography menu . Retrieved 03 14, 2011, from Chemguide: http://www.chemguide.co.uk/analysis/chromatogrmenu.html#top McKay, P. (2011, 03 14). An Introduction to Chromatography. Retrieved 03 14, 2011, from http://www.accessexcellence.org: http://www.accessexcellence.org/LC/SS/chromatography_background.php McMurry, J. (2008). Organic Chemistry. In J. McMurry, organic Chemistry (pp. 431-433). London: Thompson Brooks/Cole. Turner, P. M. (2006). Bios Instant Notes. In U. o. School of Biological sciences, Molecular Biology (p. 106). Taylor & Francis group. Unknown. (2011, march 22). BIO 102 - Biological Sciences. Retrieved march 22, 2011, from http://pages.cabrini.edu/ddunbar/BIO%20102/bio_102_-_biological_sciences.htm Unknown. (2011, march 10). Biology. Retrieved March 10, 2011, from University of Georgia: WWW.PLANTBIO.UGA.EDU E-Notes.com (2011) Recombinant DNA Available at: http://www.enotes.com/topic/Recombinant_DNA Accessed on 7/3/2011 Weaver, R. F., (2008). Molecular biology. 4th edition. New York London: McGraw-Hill Higher Education Schleif, R. F. Wensink, P. C., (1981). Practical methods in molecular biology. New York, Springer-Verlag Prasher, D., Eckenrode, V., Ward, W., Prendergast, F. and Cormier, M., 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 111: 229-33 Watson, J., (2008). Molecular biology of the gene. 6th edition, Harlow: Addison-Wesley Prasher, D., Eckenrode, V., Ward, W., Prendergast, F. and Cormier, M., 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 111: 229-33 Watson, J., (2008). Molecular biology of the gene. 6th edition, Harlow: Addison-Wesley