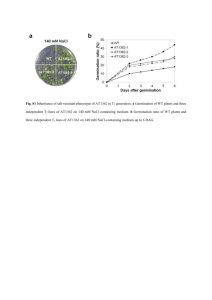
Figure S1 - Springer Static Content Server
advertisement

Supporting Information Identification of Autophagy-related Genes ATG4 and ATG8 from Wheat (Triticum aestivum L.) and Profiling of Their Expression Patterns Responding to Biotic and Abiotic Stresses Dan Pei, Wei Zhang, Hong Sun, Xiaojing Wei, Jieyu Yue, Huazhong Wang School of Life Sciences, Tianjin Normal University / Tianjin Key Laboratory of Animal and Plant Resistance, Tianjin 300387, China First author: Ms. Dan Pei School of Life Sciences, Tianjin Normal University / Tianjin Key Laboratory of Animal and Plant Resistance, Tianjin, China 393 Binshuixi Road, Tianjin 300387, China E-mail: 415017319@qq.com Corresponding author: Prof. Huazhong Wang School of Life Sciences, Tianjin Normal University / Tianjin Key Laboratory of Animal and Plant Resistance, Tianjin, China 393 Binshuixi Road, Tianjin 300387, China Telephone and fax: 86-22-23766539 E-mail: skywhz@mail.tjnu.edu.cn Journal: Plant Cell Reports Table S1. List of primers used in this paper Primer name Sequence(5' to 3') A8a/eF A8a/eR A8b/dF A8b/dR A8cF A8cR A8fF A8fR A8gF A8gR A8hF A8hR A4a/bF A4a/bR GTCGCAAACCCTCGCCCAAATCC CTCGCCCAAATCCGCCGAATCCC ACCAAGGTAGATCGATGGCGAAGA TTGATTGGCACGCCTTATTTACA GCGTCGCAAACCCTCGCCCAAAT ATAGGATAAGTCAGGATACGAATAGG CAAGGTAGATCGATGGCGAAGAG TTAAAGATCCTACGACGAGGCAA GAGCGACTTCCAATCCAGTC AATCATCCCAGAGCATGAGG TCTATCCGCGATTCAATCAAATAC CAGTCAGGCAGATGTCACCCAAA GGGTGCCTGATGCCCTGATTGAC TTCCCATCGAACCAACTTGTGCC NotI-A8a/bF ATAAGAATGCGGCCGCATGGCGAAGAGCTC GTTCAAGC NotI-A8a/bR ATAAGAATGCGGCCGCCTAGAGCAATCCGAA GGTGT NotI-A8gF ATAAGAATGCGGCCGCATGGCCAAGACTTGC TTCAA NotI-A8gR ATAAGAATGCGGCCGCTTAGGCGGAGCCGA AAGTGT NotI-A8hF ATAAGAATGCGGCCGCATGAAGTCCTTCAAG AAGGA NotI-A8hR ATAAGAATGCGGCCGCTCACCCAAATGTCTT CTCGC NotI-A4aF ATAAGAATGCGGCCGCATGACGAGCTTGCCT GAGAG NotI-A4aR ATAAGAATGCGGCCGCTCAGAGAATCTGCCA CTCAT NotI-A4bF ATAAGAATGCGGCCGCATGACGAGCTTGCCT GAGAG NotI-A4bR ATAAGAATGCGGCCGCTCAGAGAATCTGCCA CTCGT SmaI-A8a/bF TCCCCCGGGAATGGCGAAGAGCTCGTTCAA BamHI-A8a/bR CGGGATCCCTAGAGCAATCCGAAGGTGT XbaI-A8gF GCTCTAGAAGCCAAGACTTGCTTCAAGACC SacI-A8gR CGAGCTCTTAGGCGGAGCCGAAAGTGT XbaI -A8hF GCTCTAGAAATGAAGTCCTTCAAGAAGGA SacI -A8hR CGAGCTCTCACCCAAATGTCTTCTCGC qA8aF qA8aR qA8gF qA8gR qA8hF qA8hR qA4aF qA4aR qA4bF qA4bR qTubulin-F TTCCTGTGATCGTTGAGAAGGCTG AAGGTCGGCAGGGACAAGGT TTGTGAATAGCACCTTGCCACC GTGTTCTCGCCACTGTAAGTCATG GGAGGAGAGGGCGAATGAGTC GCATGTCACACGGAACCAGGTAC TATCACTGCAGCACTGTCCG GGTGTCGATGTTGTCGGAGG TCAGTTCAGCCGTCGAAACA GAGCCATCAAGGTCCTCCG GTGGAACTGGCTCTGGC qTubulin-R CGCTCAATGTCAAGGGA Product size (bp) Application 438 Amplification of TaATG8a, 8e full-length cDNA by RT-PCR 436 Amplification of TaATG8b, 8d full-length cDNA by RT-PCR 559 Amplification of TaATG8c full-length cDNA by RT-PCR 474 Amplification of TaATG8f full-length cDNA by RT-PCR 654 Amplification of TaATG8g full-length cDNA by RT-PCR 446 Amplification of TaATG8h full-length cDNA by RT-PCR 1724(4a) 1727(4b) Amplification of TaATG4a,4b full-length cDNA by RT-PCR 390 Construction of TaATG8a and 8b yeast or prokaryotic expression vectors 392 Construction of TaATG8g yeast or prokaryotic expression vectors 383 Construction of TaATG8h yeast or prokaryotic expression vectors 1470 Construction of TaATG4a yeast or prokaryotic expression vectors 1473 Construction of TaATG4b yeast or prokaryotic expression vectors 360 Construction of GFP-TaATG8a or -TaATG8b expression vectors 358 Construction of GFP -TaATG8g expression vector 351 Construction of GFP -TaATG8h expression vector 80 106 134 245 Quantitative RT-PCR of TaATG8a transcripts Quantitative RT-PCR of TaATG8g transcripts and amplification in Chinese spring derived aneuploid Quantitative RT-PCR of nulli-tetrasomic(NT) TaATG8h transcriptslines for chromosomal localization of TaATG8g RT-PCR of Quantitative TaATG4a transcripts 101 Quantitative RT-PCR of TaATG4b transcripts 234 Quantitative RT-PCR of the wheat β-tubulin gene transcripts Figure S1. Multiple alignment of wheat ATG8s and homologues from other species. The alignment was produced in ClustalX 2.1 software. Sequences in the alignment include the nine wheat ATG8s here identified, AER27507 (TdATG8) from Triticum dicoccoides, XP_003581707 (BdATG8a) and XP_003571383 (BdATG8b) from Brachypodium distachyon, Os07g0512200 (OsATG8a), Os04g0624000 (OsATG8b), Os08g0191600 (OsATG8c) and Os02g0529150 (OsATG8d) from Oryza sativa, ACJ73920 (ZmATG8a), ACJ73921 (ZmATG8b), ACJ73922 (ZmATG8c), ACJ73923 (ZmATG8d), and ACJ73925 (ZmATG8e) from Zea mays, At4g21980 (AtATG8a), At4g04620 (AtATG8b), At1g62040 (AtATG8c), At2g05630 (AtATG8d), At2G45170 (AtATG8e), At4g16520 (AtATG8f), At3g60640 (AtATG8g), At3g06420 (AtATG8h) and At3g15580 (AtATG8i) from Arabidopsis thaliana, ACU13796 (GmATG8a), ACU14633 (GmATG8b), ACU17086 (GmATG8c), BAH22449 (GmATG8d), ACU15101 (GmATG8e), ACU19559 (GmATG8f), ACU13862 (GmATG8g), ACU16419 (GmATG8h), BAH22448 (GmATG8i) and ACU19611 (GmATG8k) from Glycine max, NP_009216 (HsATG8/GATE16/GABARAPL2) from Human and NP_009475 (ScATG8) from Yeast. Names of wheat ATG8s are boxed. One hundred percent similarity, black; 80 to 100% similarity, dark gray; 60 to 80% similarity, light gray; <60% similarity, white. Hashes and asterisks respectively indicate conserved essential amino acids at the N-terminal microtubule binding site and at the ATG7 binding site. The C-terminal conservative glycine for PE-conjugation is indicated by an arrowhead. # # ## ## * Clade I Clade II * Clade I Clade II * Figure S2. Multiple alignment of wheat ATG4s and homologues from other species. The alignment was produced in ClustalX 2.1 software. Sequences in the alignment include the two wheat ATG4s here identified, Os03g0391000 (OsATG4a) and Os04g0682000 (OsATG4b) from Oryza sativa, ACJ73912 (ZmATG4a) and ACJ73914 (ZmATG4b) from Zea mays, At2g44140 (AtATG4a) and At3g59950 (AtATG4b) from Arabidopsis thaliana, NP_443168 (HsATG4a) from Human and NP_014176 (ScATG4) from Yeast. One hundred percent similarity, black; 80 to 100% similarity, dark gray; 60 to 80% similarity, light gray; <60% similarity, white. The conserved Peptidase_C54 domain is under thick line;Arrowheads mask the canonical catalytic triad of cysteine protease: Cys, Asp and His. TaATG4a TaATG4b OsATG4a OsATG4b ZmATG4b ZmATG4a AtATG4a AtATG4b ScATG4 HsATG4a MTSLPERGAAPPSNPTPSSSRE------GDAAVASSS-SSASEEQKKDGSPKQCKASILSSVLT----IFEPDQD-QSG-----RSGGHASGSYAWSRVLRRF-VGGGSMWRFLG-------CGKALTAGDVWFLGKCYKLSSEE MTSLPERGATPPSNPTSSSSCE------GDAAVASSSASSASEEQKKDGSPKQCKASILSSVLT----IFEPDQD-QSG-----RSGGHASGSYAWSRVLRRF-VGGGSMWRFLG-------CGKALTAADVWFLGKCYKLSSEE MTSLPGRGVSPSS---SDPLCE------GNAAPSSSS-----SSGQD---LKQLKNSILSCVFSSPFSIFEAHQD-SSAN----RSLKPHSGSYAWSRFLRRI-ACTGSMWRFLG-------ASKALTSSDVWFLGKCYKLSSEE MTSLPDRGVSSSS---SDPLCE------GNIAPCSSS-----SEQKEDCSLKQSKTSILSCVFNSPFNIFEAHQD-SSAN----KSPKSSSGSYDWSRVLRRI-VCSGSMWRFLG-------TSKVLTSSDVWFLGKCYKLSSEE MTTSPERGEAPPAPPRDSPCEDAAP---AAAVVASSS---SGSEREEDGGSRQPKASVLSGVFAPPLAIFEGQQQVSSTP-CDASSTKPPSGSYAWSRILRRF-VGSGSMWRLLG-------CARVLTSGDVWFLGKCYRVSPEE MTSLPERGESP-VPPLDSLRQDSAVTVLAAAAVASSS---AGSDRKEDSGSRQPKASILSGVFSPPFAIFEGQQQGSSSPACDARSTKSSSGSYGLSRILRRF-VGSGSMWRLLG-------CGRVLTSSDVWFLGKCYKVSPEE MKALCDRFVPQQCS--SSSKSD-------THDKSP-------LVSDSGPSDNKSKFTLWSNVFTSSSSVSQPYRESSTSG-----HKQVCTTRNGWTAFVKRVSMASGAIRRFQERVLGPNRTGLPSTTSDVWLLGVCYKISADE MKAICDRFVPSKCS--SSSTSE-------KRDISSPT----SLVSDSASSDNKSNLTLCSDVVASSSPVSQLCREASTSG-----HNPVCTTHSSWTVILKTASMASGAIRRFQDRVLGPSRTGISSSTSEIWLLGVCYKISEGE MQRWLQLWKMDLVQKVSHGVFE--------------------GSSEEPAALMNHDYIVLGEVYPE-------------------RDEESGAEQCEQDCRYRGEAVSDGFLSSLFG------REISSYTKEFLLDVQSRVNFTYRT -------------------------------------------------------------------------------------------MESVLSKYEDQITIFTDYLEEYPD------------TDELVWILGKQHLLKTEK 120 121 115 118 130 133 124 127 100 42 TaATG4a TaATG4b OsATG4a OsATG4b ZmATG4b ZmATG4a AtATG4a AtATG4b ScATG4 HsATG4a S-SSDSDSEGGHAAFLEDFSSRVWITYRKGFDVISDSKLTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRKPAQNPSDPEHTRILHLFGDSEVCAFSIHNLLQAGKSYG-LAAGSWVGP---------YAMCRAWQTLIRTNREQ S-SSDSDSEGGHAAFLEDFSSRVWITYRKGFDAISDSKLTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRKPAQSPSDPEHTRILHLFGDSEVCAFSIHNLLQAGKSYG-LAAGSWVGP---------YAMCRAWQTLIRTNREQ L-SNSSDCESGNAAFLEDFSSRIWITYRKGFDAISDSKYTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRKPSQKPYSPEYIGILHMFGDSEACAFSIHNLLQAGKSYG-LAAGSWVGP---------YAMCRAWQTLVCTNREH S-SSDSDSESGHATFLEDFSSRIWITYRRGFDAISDSKYTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRRPLEKPYNPEYIGILHMFGDSEACAFSIHNLLQAGNSYG-LAAGSWVGP---------YAMCRAWQTLVRTNREQ EESGGSDSDSGHAAFLEDFSSRIWITYRKGFDAIPGSKLTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRKPSEKPYDPDYIRVLHLFGDSEACAFSIHNLLQAGRNYG-LAAGSWVGP---------YAMCRAWQTLIRTNREQ EESGDSESDSGHAAFLEDFSSRIWITYRKGFDAISDSKLTSDVNWGCMVRSSQMLVAQALIFHHLGRSWRKPPEKPYNPDYIGVLHLFGDSEACAFSIHNLLQAGRNYG-LAAGSWLGP---------YAMCRAWQTLIRTNREQ N-SGETDTGTVLAALQLDFSSKILMTYRKGFEPFRDTTYTSDVNWGCMIRSSQMLFAQALLFHRLGRAWTKK-SELPEQEYLETLEPFGDSEPSAFSIHNLIIAGASYG-LAAGSWVGP---------YAICRAWESLACKKRKQ S-SEEADAGRVLAAFRQDFSSLILMTYRRGFEPIGDTTYTSDVNWGCMLRSGQMLFAQALLFQRLGRSWRKKDSEPADEKYLEILELFGDTEASAFSIHNLILAGESYG-LAAGSWVGP---------YAVCRSWESLARKNKEE RFVPIARAPDGPSPLSLNLLVRTNPISTIEDYIANPDCFNTDIGWGCMIRTGQSLLGNALQILHLGRDFRVNGNESLERE-SKFVNWFNDTPEAPFSLHNFVSAGTELSDKRPGEWFGP---------AATARSIQSLIYG---S------------KLLSDISARLWFTYRRKFSPIGGTGPSSDAGWGCMLRCGQMMLAQALICRHLGRDWSWEKQKEQPKEYQRILQCFLDRKDCCYSIHQMAQMGVGEG-KSIGEWFGPNTVAQVLKKLALFDEWNSLAVYVSMD 254 255 249 252 265 268 257 261 231 174 TaATG4a TaATG4b OsATG4a OsATG4b ZmATG4b ZmATG4a AtATG4a AtATG4b ScATG4 HsATG4a PEVINRNECFPMALYVVSGDEDGERGGAPVVCIDVAAQLCYDFNKDQSTWSPILLLVPLVLGLDKINPRYIPLLKETFTFPQSLGILGGKPGASTYIAGVQDDRALYLDPHEVQLAVNIASDNLEANTSSYHCSTVRDMPLDLID PEVINRNESFPMALYVVSGDEDGERGGAPVVCIDVAAQLCYDFNKDQSAWSPILLLVPLVLGLDKINPRYIPLLKETFTFPQSLGILGGKPGASTYIAGVQDDRALYLDPHEVQMAVNIASDNLEADTSSYHCSTVRDMPLDLID HEAVDGNGNFPMALYVVSGDEDGERGGAPVVCIDVAAQLCCDFNKNQSTWSPILLLVPLVLGLDKLNPRYIPLLKETLTFPQSLGILGGKPGTSTYIAGVQDDRALYLDPHEVQLAVDIAADNLEAGTSSYHCSTVRDLALDLID HEVVDGNESFPMALYVVSGDEDGERGGAPVVCIDVAAQLCCDFNKGQSTWSPILLLVPLVLGLDKINPRYIPLLKETFTFPQSLGILGGKPGTSTYIAGVQDDRALYLDPHEVQMAVDIAADNIEADTSSYHCSTVRDLALDLID ADAVDGKENFPMALYVVSGDEDGERGGAPVFCIDVAAQLCSNFNKGQCTWSPILLLIPLVLGLDKINPRYIPLLKETFKFPQSLGILGGKPGTSTYIAGVQEDRALYLDPHDVQMAVDIAPDNLEADTSSYHCSVVRDLALEQID ADAVDGKENFPMALYVVSGDEDGERGGAPVVCIDVAAQLCSDFNKGPSTWSPILLLVPLVLGLDKINPRYIPLLKETFMFPQSLGILGGKPGTSTYIAGVQDDRALYLDPHEVQMTVDIALDNLEADTSSYHCSVVRALALEQID TD--SKNQTLPMAVHIVSGSEDGERGGAPILCIEDATKSCLEFSKGQSEWTPIILLVPLVLGLDSVNPRYIPSLVATFTFPQSVGILGGKPGASTYIVGVQEDKGFYLDPHEVQQVVTVNKETPDVDTSSYHCNVLRYVPLESLD TD--DKHKSFSMAVHIVSGSEDGERGGAPILCIEDVTKTCLEFSEGETEWPPILLLVPLVLGLDRVNPRYIPSLIATFTFPQSLGILGGKPGASTYIVGVQEDKGFYLDPHDVQQVVTVKKENQDVDTSSYHCNTLRYVPLESLD ----FPECGIDDCIVSVSSGDIYENEVEKVFAENPN--------------SRILFLLGVKLGINAVNESYRESICGILSSTQSVGIAGGRPSSSLYFFGYQGNEFLHFDPHIPQPAVEDS------FVESCHTSKFGKLQLSEMD NTVVIEDIKKMCRVLPLSADTAGDRPPDSLTASNQSKGTSAYCS----AWKPLLLIVPLRLGINQINPVYVDAFKECFKMPQSLGALGGKPNNAYYFIGFLGDELIFLDPHTTQTFVDTEENGTVNDQTFHCLQSPQRMNILNLD TaATG4a TaATG4b OsATG4a OsATG4b ZmATG4b ZmATG4a AtATG4a AtATG4b ScATG4 HsATG4a PSLAIGFYCRDKDDFDDFCSRASE------LAEQANGAPLFTVVQSVQPSKQMYNQDDGSGCSGY---------------GASDNIDTEDLDGSGETGEDEWQIL------------------------------------PSLAIGFYCRDKDDFDDFCSRASE------LAEQANGAPLFTVVQSVQPSKQMYNQDDGSGCSGY---------------GVSDNIDAEDLDGSGETGEDEWQIL------------------------------------PSLAIGFYCRDKDDFDDFCSRASE------LVDKANGAPLFTVVQSVQPSKQMYNEESSSGD-------------------GMDSINVEGLDGSGETGEEEWQIL------------------------------------PSLAIGFYCRDKDDFDDFCSRATE------LVDKANGAPLFTVVQSVQPSKQMYNQDDVLGIS------------------GDGNINVEDLDASGETGEEEWQIL------------------------------------PSLAIGFYCRDKDDFDDFCSRASE------LAEKANGAPLFTVMQSVQPSKQMYKQDDGLCCCSG---------------SSMANEDY-DLDASGEAG-EEWQIL------------------------------------PSLAIGFYCRDKDDFDDFCSRASE------LAEKANGAPLFTVVQSIEPSKQMYKQDDGLG-CSG---------------SSMANDD--DLDGSGEA--EEWQIL------------------------------------PSLALGFYCRDKDDFDDFCLRALK------LAEESNGAPLFTVTQTHT------AINQSNY--------------------GFAD------DDSEDEREDDWQML------------------------------------PSLALGFYCQHKDDFDDFCIRATK------LAGDSNGAPLFTVTQSHRRNDCGIAETSSST--------------------ETST------EISGEEHEDDWQLL------------------------------------PSMLIGILIKGEKDWQQWKLEVAESAIINVLAKRMDDFDVSCSMDDVESVSSNSMKKDASNNENLGVLEGDYVDIGAIFPHTTNTEDVDEYDCFQDIHCKKQKIVVMGNTHTVNANLTDYEVEGVLVEKETVGIHSPIDEKC PSVALGFFCKEEKDFDNWCSLVQK------EILKEN-LRMFELVQKHPSHWPPFVPPAKPEVTTT---------------GAEFIDSTEQLEEFDLEEDFEILSV------------------------------------- Peptidase_C54 domain 399 400 394 397 410 413 400 404 352 315 483 484 474 478 492 492 467 477 494 398