Journal name - Springer Static Content Server
advertisement

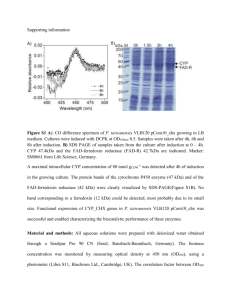
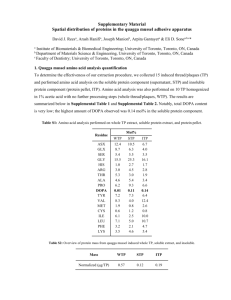
Journal name: Amino Acids Supplementary Information Employing in vitro analysis to test the potency of methylglyoxal in inducing the formation of amyloid like aggregates of caprine brain cystatin Waseem Feeroze Bhat, Sheraz Ahmad Bhat, Peerzada Shariq Shaheen Khaki, Bilqees Bano* Department of Biochemistry, Faculty of Life Sciences, Aligarh Muslim University, Aligarh 202002, UP, India *Corresponding author: Prof. Bilqees Bano Phone: (+91) 0571-2700920 (3721, 3720) Email address: bilqeesbano99@gmail.com Molecular Weight Determination (a) Molecular weight determination by gel filtration. Molecular weight of Cystatin in native condition was determined by using gel filtration chromatography on sephacryl S-100HR column (Fig.3b). The marker proteins were A, BSA (66 kDa); B, Ovalbumin (44.3 kDa); C, Trypsin (23.3 kDa); D, Lysozyme (14.3 kDa) E, Cytochrome C (12.3 kDa); the arrow shows position of CBC. These proteins were chromatographed after equilibration of the column sephacryl S-100HR with 0.05 M sodium phosphate buffer pH 7.5 and the elution volume was determined by passing them from column. Analysis of the data indicated linear relationship between log (kDa) and Kav by the method of Andrews (1965). Where Ve is the elution volume and Vo is the void volume of the column. The native cystatin corresponds to molecular weight of ~ 44 kDa. Fig.3b. Plot of log (kDa) vs Kav for markers passed through Sephacryl S 100HR column: A, BSA (66 kDa); B, Ovalbumin (44.3 kDa); C, Trypsin (23.3 kDa); D, Lysozyme (14.3 kDa) E, Cytochrome C (12.3 kDa); the arrow shows position of CBC. (b) Molecular weight determination by SDS PAGE REDUCING AND NONREDUCING SDS PAGE Purified cystatin was analyzed by SDS-PAGE under reducing conditions by the method of Weber and Osborn (1969), (in the presence of β-mercaptoethanol) and non reducing conditions (in the absence of β-mercaptoethanol ) Fig.3c, Brain Cystatin migrated as two bands with different motilities in both the conditions suggesting a two subunit structure which are held together by non-covalent forces. Fig.3c. Molecular weight determination of cystatin by SDS-PAGE electrophoresis on 12.5% polyacrylamide gel at 25oC.Middle lane contained the molecular mass standards: A, Phosphorylase b (97.4 kDa); B, Bovine serum albumin (66 kDa); C, Ovalbumin (44.3 kDa); D, Carbonic anhydrase (29.1 kDa); E, Soyabean trypsin inhibitor (20.1 kDa); F, Lysozyme (14.3 kDa). Lane a contained 40 µg protein without β-mercaptoethanol and lane b contained 40 µg of β-mercaptoethanol treated purified enzyme. . The molecular weight of CBC was determined under denaturing conditions by the method of Weber and Osborn (1969). The relative Rf vlue of each marker protein was determined. The markers used were 1, Phosphorylase b 2, BSA 3, Oval albumin 4, Carbonic anhydrase 5, Soyabean trypsin inhibitor 6, Lysozyme, was plotted against log molecular weight (Fig.3d) positions of two subunit corresponding to molecular weight of 30.19 kDa and another of 13.8 kDa were obtained. The least square analysis of the data indicated a linear relationship log M and relative mobility which gave the molecular weight ~ 44 kDa. Fig.3d. Log (kDa) of marker proteins was plotted against Rf of molecular weight markers for determination of molecular mass of brain cystatin. Molecular weight markers of standard proteins are A, Phosphorylase b (97.4 kDa); B, Bovine serum albumin (66 kDa); C, Ovalbumin (44.3 kDa); D, Carbonic anhydrase (29.1 kDa); E, Soyabean trypsin inhibitor (20.1 kDa); F, Lysozyme (14.3 kDa). The molecular weight for the brain cystatin subunits were determined as, Subunit-I (30.19 kDa), and Subunit-II (13.8 kDa). Andrews P (1965) The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J 96:595-606 Weber K, Osborn M (1969) The reliability of molecular weight determinations by dodecyl sulfatepolyacrylamide gel electrophoresis. J Biol Chem 244:4406-12.