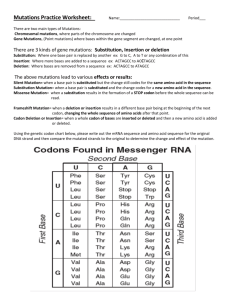
Instructions for CAN1 gene analysis.
advertisement

Luria Delbruck Experiment Goal: To analyze CAN1 sequences from surviving yeast cells after plating on canavanine (both wild type and mutator strains). We'd like to see which mutations are non-synonymous. 1- Open Geneious. 2- Import CAN1 sequences and CAN1 reference sequence. 3- Select 5 contigs from each CAN1 sequence group and the reference CAN1 sequence. 4- Choose "Map to reference" from the Align/Assemble folder. CAN1 reference sequence (because it's longer than each contig) will have already been selected as reference. Click ok. 5- When looking for mutations, look for majority consensus...Also avoid counting mutations from sequence regions containing "N", particularly if they don't agree with other contings. 6- When you find a mutation, or an indel (which you will see as "-"), write down the number of bp at which it occurs with respect to the reference CAN1 gene (this is important). 7- Now translate the reference CAN1, and choose frame 1. This is the only frame with a terminating codon at the end and starter codon in the beginning of the sequence. 8- To see if the mutation or indel were non-synonymous, first create a copy of the reference CAN1 gene, turn on editing function, and make the mutation/indel you observed. 9- Now translate the above sequence (frame 1), and do a protein alignment with the translated reference CAN1 gene. Do you see a change between these two sequences?