1-31 oct - WordPress.com
advertisement

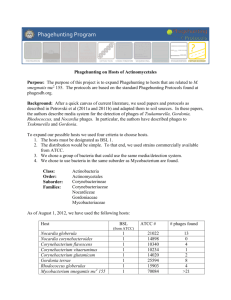
DATE- 1-31 October DNA Extraction by PCI (phenol cholorofom isoamylalcohol)/SDS Method PROCEDURE 1. 1 ml of lysate was transferred to a 15ml tube 2. 12.5ul 1M MgCl2 was added and mixed gently. 3. 0.8ul DNAse (1000U/ml) and 2ul RNASE A (50 mg/ml) was added to the lysate-MgCl2 mixture and briefly vortexed and incubated at room temperature for 30 mins. 4. The following reagents/enzymes were added in the order listed below a) 40ul of 0.5M EDTA b) 2.5ul of Proteinase K (20mg/ml) c) 50ul of 10% SDS 5. The mixture was vortexed vigorously. 6. Incubated at 550C or 60 mins. Vortexed the mixture vigorously twice during incubation at intervals of 20mins. 7. Transferred 500ul of the mixture to 1.5ml microcentrifuge tube. 8. Transferred 500ul PCI to each tube with 500ul tube. 9. Centrifuged for 5 mins at room temperature at 13K rpm. 10. The top aqueous layer was removed above the interphase 11. Steps 8 and 9 were repeated till the white interphase was gone. 12. Precipitate the DNA a) Added 1ml of 95% ethanol and 50ul of 3M sodium acetate solution to the top aqueous layer b) Placed the sample for on ice for 5 mins 13. Centrifuged at room temperature at 13K for 10 mins. 14. The tubes were decanted carefully to not to lose the pellet. Then 500ul of 70% ethanol was added to wash the pellet. Ethanol was just allowed to run through the pellet. 15. Centrifuged for 10 mins at 13K rpm at room temperature 16. The tubes were decanted. 17. Air dried the pellet till the smell of ethanol was gone. 18. Dissolved DNA in 10ul of sterile distilled water. To ensure complete solvation the tubes were set at 4 oC overnight Agarose gel analysis (0.8%) of three phage DNA To study the growth curve of M.smegmatis mc2 155 was studied 1. Streaking was done from the glycerol stock on M7H10/CHX/CB plate and incubated at 370C for three days. 2. A single colony was picked up and inoculated in 5ml M7H9/CHX/CB Tween media and grown till 24 hrs. 3. Optical density (O.D.) at 600nm was taken at intervals of 3hrs. O.D. for blank was 0.039 Plotted absorbance of the culture (difference in table below) was adjusted with the blank Time 0 6 9 12 24 30 33 36 48 54 57 60 72 Abs1 0 0.05 0.073 0.136 0.649 0.778 0.813 0.875 0.879 0.834 0.778 0.768 0.562 0.9 Abs2 0 0.051 0.077 0.146 0.687 0.869 0.845 0.874 0.771 0.857 0.869 0.778 0.6 Mean Diff 0 0 0.0505 0.0105 0.075 0.041 0.141 0.107 0.668 0.645 0.8235 0.7855 0.829 0.8005 0.8745 0.834 0.825 0.7825 0.8455 0.813 0.8235 0.7855 0.773 0.735 0.581 0.543 Growth curve of M.smegmatis mc2 155 Absorbance (600nm) 0.8 0.7 0.6 0.5 0.4 0.3 0.2 0.1 0 0 3 6 9 12 15 18 21 24 27 30 33 36 39 42 45 48 51 54 57 60 63 66 69 72 75 78 Time (hr) Secondary inoculation (for infection) was done in 50 ml of M7H9/CHX/CB Tweenless media at 20-24hrs of time interval Screening of soil samples ISOLATION OF MYCOBACTERIOPHAGES FROM ENVIRONMENTAL SAMPLES. I. Sample Collection and Preparation 1. Samples were collected in 15-50 ml conical tubes. -Liquid or solid samples ie water and soil samples were collected . - In case of solid sample, phage buffer was added with 0.1M CaCl 2 and vortexed well and kept for 15- 20 mins which allows phages to diffuse into buffer . 2. Filter sterilization of the sample -After the sample settled at the bottom of tube, 1 ml of the sample was drawn from the top liquid layer of phage buffer into a sterile 1.5 ml microfuge tube and filter sterelized by passing it through a 0.22 μ filter attached to a syringe. II. First Round of Infection: Plaque Screening 1. Infect 0.5 ml Mycobacterium smegmatis mc2155 with 50 μl of filtered sample In sterile tubes (one for each sample plus one for a negative control), 0.5 ml was of M. smegmatis mc2155 was added. To each 0.5 ml aliquot of M. smegmatis, 50 μl of the filtered sample was added to infect the bacteria and allowed to stand for 30 mins. 2. Add 4.5 ml top agar with 0.1M CaCl2 Top Agar was completely melted and cooled enough.Then 4.5 ml MBTA top agar with 0.1M CaCl 2 was added to each tube of infected bacteria. 3. Plate total volume of 5.05 ml (4.5ml top agar + 0.5 ml M. smegmatis + 50μl of filtered sample) on 7H10/CB/CHX The cells and top agar mix was added on to the agar plates and each plate was swirled to evenly spread the top agar before it cooled. 4. Incubate at 37°C overnight After the samples hardened , the plates were kept upside-down at 37°C to prevent liquid condensation drops from forming on the lid and dripping onto the plate. 5. Checking of plates for plaque The plates were checked for the presence of plaques after 24-48 hrs of incubation. If there were no plaques, new samples were further collected and the procedure repeated. LOCATION MYCOBACTERIOPHAGE Chennai - Kolkata - Canada - Mahabalipuram - Faridabad 2 - Faridabad 3 - Noida 1 - Noida 2 - Noida 3 - Ghaziabad 1 - Ghaziabad 2 - Ghaziabad 3 - SUBMISSION OF MYCOBACTERIOPHAGES TO THE DATABASE All the Mycobacteriophages purified and amplified have been submitted to the phage database maintained by University Of Pittsburgh, USA The Mycobacteriophage Database