Supplementary Table 1 Primers used for quantitative PCR
advertisement

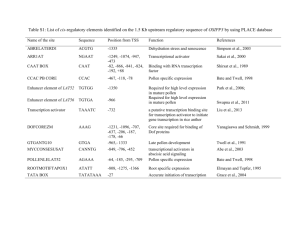
Supplementary Table 1 Primers used for quantitative PCR Gene Forward primer Reverse primer OsFER AGGGATTCGCCAAATTCTTC CCTTTCTCAGGATGGTCGAA NAS1 AGCGCGGTTGAGAAGGCAGAA GGCATGTTCCTCGTTTACACTGGC NAS2 CGCCAAGAGGCTATCCGTCTGA GATCAATGCCCAGGCGCCTA NAS3 CGAGTGTTCGACTGATCACC CCGGTGATCTTCTCAACCAA NAAT1 TGCTGTGGCAGATCATTTGT TTGGGTAGCCTGGTCTAGGA SoyFERH1 GGGATTTGCCAAGTTCTTCA TACAATGCATCCCCCTTTTC YSL15 TGATCATAGCCTACGCGCTCGG GCGATGAAGCAGCCAATGCC YSL18 GACAAGCGAAGAGGCAAGTC CGATTCCCACATAAGTTCCG FRO1 GGCCACCAAGCAGTCCCTCTTT CGAAAGGCCCGTCCACAAGA FRO2 TCCCTGTTGCCGCTCATTGG CCCGGGGACGTTTGCAACTT Supplementary Table 2 Grain quality assessment of transgenic IR64 lines Sample ID DESIGNATION IR64 control Cooking traits AC GT GC Low HI/I Soft PC (%) 12.2 Low I Soft Intermediate I Intermediate HI/I Milling potential score DB C 1 1 11.6 1 1 Soft 12.5 1 2 Soft 12.6 1 2 Single transformation IR64-GLUB4:: SoyFERH1018B-003 IR64- GLUB4::OsFER1C-088A006 IR64-GLUB1:: SoyFERH2176A-002 Co-transformation IR64- GLUB1:: SoyFERH1Low HI/I Soft 12.5 1 2 028A-109 IR64- GLUB1:: SoyFERH1Intermediate HI/I Soft 10.1 1 2 072A-094 IR64-GLUB1:: SoyFERH1-37A- Intermediate I Soft 11.7 1 2 129 AC = amylose content: waxy (0-2%), very low (3-9%), low (10-19%), intermediate (20-25%), high (>25%); GT = gelatinization temperature: I = intermediate (70-72 C), HI = high intermediate (73-74 C), results are from six replicates; GC = gel consistency scored as 1) very flaky rice with hard gel consistency (length of gel, 40 mm or less); 2) flaky rice with medium gel consistency (length of gel, 41 to 60 mm); and 3) soft rice with soft gel consistency (length of gel, more than 61 mm), results are from two replicates; PC = protein content, with range of 8 to 14 considered as normal or marketable; DB = degree of breakage and C = chalkiness score, both classified using the following scale: 1 = mostly whole translucent grains; 2 = few broken/chalky grains, and 3 = >70% of grains are broken; >50% chalky grains Supplementary Table 3 Comparison of average iron concentration of IR64 transgenic plants (single transformants) with ferritin genes driven by GLUB1 versus GLUB4 promoter. Average iron concentration for each promoter were obtained from 8 transgenic plants Promoter T2 ICP (mg kg-1) T3 ICP (mg kg-1) GLUB1 5.63 ± 0.67 4.60 ± 1.20 GLUB4 6.21 ± 0.74 5.03 ± 1.80 IR64:GLUB1:GUS IR64:GLUB4:GUS IR64:GLB:GUS IR64:35S:GUS IR64 Dough (15 DAF) Mature (30 DAF) Supplementary Fig 1 Histochemical GUS staining of dough (left) and mature seeds (right) of transgenic IR64 plants with different promoter:GUS constructs M 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 WT kb 8.5 7.4 6.1 4.8 3.6 2.7 Supplementary Fig. 2 Southern blot analysis of transgenic IR64 with SoyFERH1 driven by GLUB1 promoter. Genomic DNA from each sample digested with EcoRI, hybridized with a PCR-labeled probe containing 928-bp fragment of SoyFERH1 gene. M = Dig-labeled marker VII, WT = wild-type IR64, 1-28 = T0 events with SoyFERH1gene Supplementary Fig. 3 Screenhouse evaluation of phenotype of different T1 transgenic lines Fe mg kg -1 9 8 7 6 5 4 3 2 1 0 T2 ICP T3 ICP R FE s O 1C R FE y So H1 R FE y So H2 Ferritin Gene Supplementary Fig. 4 Comparison of iron concentration of IR64 transgenic plants with different ferritin genes [GENETYX-MAC: Multiple Alignment] Date : 2013.03.08 Transit peptide OsFER1 1 -----MLPPRVAPAAAAAAPTYLAAAASTPASVWLPVPRGAGPGAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD [GENETYX-MAC: Multiple Alignment] OsFER2 1 -MLPPRVAPSSLAAAAAAAPTYLAAAASTPASVWLPVPRGAGAVAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD Date : 2013.03.08 SoyFERH1 1 MALAPSKVSTFSGFSPKPSVGGAQKNPTCSVSLSFLNEKLGSRNLRVCAST---VPLTGVIFEPFEEVKKSELAVPTAPQVSLARQNYAD SoyFERH2 1 MALSCSKVLSFY-LSPVVGGGDVPKKLTFSSFLGLSKGVGGSRSSRVCAASNAPAPLAGVIFEPFQELKKDYLAVPIAHNVXLARQNYAD [GENETYX-MAC: Multiple Alignment] SoyFERH3 1 --MLLRTASSFSLLKANADHILPLPNSSSSGIIRYSQSLGKNLVPCATKD-TNNRPLTGVVFEPFEEVKKELDLVPTVPQASLARQKYTD OsFER1 1 -----MLPPRVAPAAAAAAPTYLAAAASTPASVWLPVPRGAGPGAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD Date : 2013.03.08 SoyFERH4 1 -----------MLLRTAAASASSLSLFSPNAEPPRSVP-ARGLVVRAAKGSTNHRALTGVIFEPFEEVKKELDLVPTVPQASLARQKYVD OsFER2 1 -MLPPRVAPSSLAAAAAAAPTYLAAAASTPASVWLPVPRGAGAVAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD Consensus 1 --------------------------------------------------------LXGVXFZPFZEXKXXXXXVPXXXXXXLARQXXXD SoyFERH1 1 MALAPSKVSTFSGFSPKPSVGGAQKNPTCSVSLSFLNEKLGSRNLRVCAST---VPLTGVIFEPFEEVKKSELAVPTAPQVSLARQNYAD SoyFERH2 1 MALSCSKVLSFY-LSPVVGGGDVPKKLTFSSFLGLSKGVGGSRSSRVCAASNAPAPLAGVIFEPFQELKKDYLAVPIAHNVXLARQNYAD OsFER1 1 -----MLPPRVAPAAAAAAPTYLAAAASTPASVWLPVPRGAGPGAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD 86 ECEAAINEQINVEYNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLIKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYGY SoyFERH3 1 --MLLRTASSFSLLKANADHILPLPNSSSSGIIRYSQSLGKNLVPCATKD-TNNRPLTGVVFEPFEEVKKELDLVPTVPQASLARQKYTD OsFER2 1 -MLPPRVAPSSLAAAAAAAPTYLAAAASTPASVWLPVPRGAGAVAVCRAAGKGKEVLSGVVFQPFEELKGELSLVPQAKDQSLARQKFVD 90 ECEAAISEQINVEFNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLMKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYAM SoyFERH4 1 -----------MLLRTAAASASSLSLFSPNAEPPRSVP-ARGLVVRAAKGSTNHRALTGVIFEPFEEVKKELDLVPTVPQASLARQKYVD SoyFERH1 1 MALAPSKVSTFSGFSPKPSVGGAQKNPTCSVSLSFLNEKLGSRNLRVCAST---VPLTGVIFEPFEEVKKSELAVPTAPQVSLARQNYAD 88 ECESAINEQINVEYNASYVYHSLFAYFDRDNVALKGFAKFFKESSEEEREHAEKLMKYQNTRGGRVVLHPIKNAPSEFEHVEKGDALYAM Consensus 1 --------------------------------------------------------LXGVXFZPFZEXKXXXXXVPXXXXXXLARQXXXD SoyFERH2 1 MALSCSKVLSFY-LSPVVGGGDVPKKLTFSSFLGLSKGVGGSRSSRVCAASNAPAPLAGVIFEPFQELKKDYLAVPIAHNVXLARQNYAD 90 DSESAINEQINVEYNVSYVYHALFAYFDRDNIALKGLAKFFKESSEEEREHAEQLIKYQNIRGGRVVLHPITSPPSEFEHSEKGDALYAM SoyFERH3 1 --MLLRTASSFSLLKANADHILPLPNSSSSGIIRYSQSLGKNLVPCATKD-TNNRPLTGVVFEPFEEVKKELDLVPTVPQASLARQKYTD 88 DCEATINEQINVEYNVSYVYHAMFAYFDRDNVALKGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSEFDHEEKGDALYAM OsFER1 86 ECEAAINEQINVEYNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLIKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYGY SoyFERH4 1 -----------MLLRTAAASASSLSLFSPNAEPPRSVP-ARGLVVRAAKGSTNHRALTGVIFEPFEEVKKELDLVPTVPQASLARQKYVD 79 ESESAVNEQINVEYNVSYVYHAMFAYFDRDNVALRGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSDFDHADKGDALHAM OsFER2 90 ECEAAISEQINVEFNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLMKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYAM Consensus 1 --------------------------------------------------------LXGVXFZPFZEXKXXXXXVPXXXXXXLARQXXXD 35 XXEXXXXEQINVEXNXSYXYHXXFAYFDRDNXALXGXAKFFKESSXEERXHAEXLXXYQNXRGGXVXLXXIXXXXXXFXHXXKGDALXXX SoyFERH1 88 ECESAINEQINVEYNASYVYHSLFAYFDRDNVALKGFAKFFKESSEEEREHAEKLMKYQNTRGGRVVLHPIKNAPSEFEHVEKGDALYAM SoyFERH2 90 DSESAINEQINVEYNVSYVYHALFAYFDRDNIALKGLAKFFKESSEEEREHAEQLIKYQNIRGGRVVLHPITSPPSEFEHSEKGDALYAM OsFER1 86 ECEAAINEQINVEYNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLIKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYGY 176 FVL-----------------------------------------------------------------------------------SoyFERH3 88 DCEATINEQINVEYNVSYVYHAMFAYFDRDNVALKGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSEFDHEEKGDALYAM OsFER2 90 ECEAAISEQINVEFNASYAYHSLFAYFDRDNVALKGFAKFFKESSDEERDHAEKLMKYQNMRGGRVRLQSIVTPLTEFDHPEKGDALYAM 180 ELALALEKLVNEKLHNLHSVASRCNDPQLTDFVESEFLEEQVKNLLPTWVEAIKKISEYVAQLRRVGKGHGVWHFDQKLLEEEA--SoyFERH4 79 ESESAVNEQINVEYNVSYVYHAMFAYFDRDNVALRGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSDFDHADKGDALHAM SoyFERH1 88 178 ECESAINEQINVEYNASYVYHSLFAYFDRDNVALKGFAKFFKESSEEEREHAEKLMKYQNTRGGRVVLHPIKNAPSEFEHVEKGDALYAM ELALSLEKLVNEKLLNVHSVADRNNDPQMADFIESEFLSEQV--------ESIKKISEYVAQLRRVGKGHGVWHFDQRLLD-----Consensus 35 XXEXXXXEQINVEXNXSYXYHXXFAYFDRDNXALXGXAKFFKESSXEERXHAEXLXXYQNXRGGXVXLXXIXXXXXXFXHXXKGDALXXX SoyFERH2 90 DSESAINEQINVEYNVSYVYHALFAYFDRDNIALKGLAKFFKESSEEEREHAEQLIKYQNIRGGRVVLHPITSPPSEFEHSEKGDALYAM 180 ELALSLEKLTNEKLLHVHSVAERNNDPQXADFIESEFLYEQV--------KSIKKIAEYVAQLRLVGKGHGVWHFDQKLLHDEDHVSoyFERH3 88 DCEATINEQINVEYNVSYVYHAMFAYFDRDNVALKGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSEFDHEEKGDALYAM 178 ELALSLEKLTNEKLLNLHSVASKNNDVQLADFIESEFLGEQV--------EAIKKISEYVAQLRRVGKGHGVWHFDQMLLHEEGVAA OsFER1 176 FVL-----------------------------------------------------------------------------------SoyFERH4 79 ESESAVNEQINVEYNVSYVYHAMFAYFDRDNVALRGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSDFDHADKGDALHAM 169 ELALSLEKLTNEKLLNLHSVATKNGDVQLADFVETEYLGEQV--------EAIKRISEYVAQLRRVGKGHGVWHFDQMLLHEGGDAA OsFER2 180 ELALALEKLVNEKLHNLHSVASRCNDPQLTDFVESEFLEEQVKNLLPTWVEAIKKISEYVAQLRRVGKGHGVWHFDQKLLEEEA--Consensus 35 XXEXXXXEQINVEXNXSYXYHXXFAYFDRDNXALXGXAKFFKESSXEERXHAEXLXXYQNXRGGXVXLXXIXXXXXXFXHXXKGDALXXX 125 XXXLXLEKLXNEKLXXXHSVAXXXXDXQXXDFXEXEXLXEQVKNLLPTWVXXIKXIXEYVAQLRXVGKGHGVWHFDQXLLXXXXXXA SoyFERH1 178 ELALSLEKLVNEKLLNVHSVADRNNDPQMADFIESEFLSEQV--------ESIKKISEYVAQLRRVGKGHGVWHFDQRLLD-----SoyFERH2 180 ELALSLEKLTNEKLLHVHSVAERNNDPQXADFIESEFLYEQV--------KSIKKIAEYVAQLRLVGKGHGVWHFDQKLLHDEDHVOsFER1 176 FVL-----------------------------------------------------------------------------------SoyFERH3 178 ELALSLEKLTNEKLLNLHSVASKNNDVQLADFIESEFLGEQV--------EAIKKISEYVAQLRRVGKGHGVWHFDQMLLHEEGVAA OsFER2 180 ELALALEKLVNEKLHNLHSVASRCNDPQLTDFVESEFLEEQVKNLLPTWVEAIKKISEYVAQLRRVGKGHGVWHFDQKLLEEEA--SoyFERH4 169 ELALSLEKLTNEKLLNLHSVATKNGDVQLADFVETEYLGEQV--------EAIKRISEYVAQLRRVGKGHGVWHFDQMLLHEGGDAA SoyFERH1 178 ELALSLEKLVNEKLLNVHSVADRNNDPQMADFIESEFLSEQV--------ESIKKISEYVAQLRRVGKGHGVWHFDQRLLD-----Consensus 125 XXXLXLEKLXNEKLXXXHSVAXXXXDXQXXDFXEXEXLXEQVKNLLPTWVXXIKXIXEYVAQLRXVGKGHGVWHFDQXLLXXXXXXA SoyFERH2 180 ELALSLEKLTNEKLLHVHSVAERNNDPQXADFIESEFLYEQV--------KSIKKIAEYVAQLRLVGKGHGVWHFDQKLLHDEDHVSoyFERH3 178 ELALSLEKLTNEKLLNLHSVASKNNDVQLADFIESEFLGEQV--------EAIKKISEYVAQLRRVGKGHGVWHFDQMLLHEEGVAA SoyFERH4 169 ELALSLEKLTNEKLLNLHSVATKNGDVQLADFVETEYLGEQV--------EAIKRISEYVAQLRRVGKGHGVWHFDQMLLHEGGDAA Consensus 125 XXXLXLEKLXNEKLXXXHSVAXXXXDXQXXDFXEXEXLXEQVKNLLPTWVXXIKXIXEYVAQLRXVGKGHGVWHFDQXLLXXXXXXA helix - helix C- helix D- helix F- helix Supplementary Fig. 5 In silico analysis o f amino acid sequence of ferritin from soybean and rice 85 89 87 89 87 85 78 89 34 87 89 85 175 87 89 179 78 87 177 34 89 179 87 177 175 78 168 179 34 124 177 179 175 178 177 179 263 168 177 250 124 179 257 177 256 178 168 247 263 124 211 250 257 178 256 263 247 250 211 257 256 247 211