Supplementary Information (docx 1010K)
advertisement

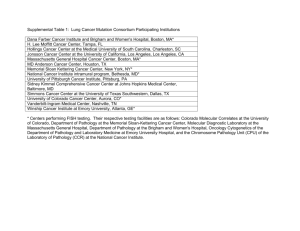
1 Supplementary Material 2 Contents 3 Supplementary Materials and Methods 4 Supplementary Figures 1 and 2 5 Supplementary Tables 1, 2, 3 and 4 6 Supplementary References 7 8 Supplementary Material and Methods 9 Patients and samples 10 Between 04/1995 and 07/2012, a total of 438 pediatric T-LBL patients were registered in the 11 NHL-BFM study center for the trials NHL-BFM95 and EURO-LB02 after informed consent. 12 The accompanying molecular research was approved by the ethical committees of Hannover 13 Medical School and Justus Liebig University Giessen, Germany. Diagnosis of T-LBL was 14 made according to WHO Classification of Hematological Malignancies and European 15 childhood lymphoma pathology panel.1 Patients were treated according to an ALL-BFM-type 16 treatment strategy for LBL as described previously.2 Data on patients characteristics, 17 diagnostics, treatment, and outcome were obtained from the clinical trial databases and 18 correlated with experimental results. 19 PCR amplification of genomic sequences and Sanger sequencing 20 A total of 114 DNA samples of pediatric T-LBL patients were used for validation of PIK3R1, 21 PIK3CA, and PTEN using Sanger sequencing, and 99 DNA samples were available for NRAS, 22 and KRAS validation. The isolation of DNA from lymphoma material has been described 23 earlier.3 24 Mutational analysis was carried out by Sanger sequencing of an appropriate genomic region 25 of the genes PIK3R1, PIK3CA, PTEN, NRAS, and KRAS after PCR amplification. For 26 amplification of PIK3R1, PIK3CA, PTEN sequences, Q5 High Fidelity DNA Polymerase was 27 used, for amplification of NRAS and KRAS, we used OneTaq DNA polymerase (New 28 England Biolabs, Frankfurt/Main, Germany) using standard buffers and conditions. 29 Amplification of KRAS exon 2 required using GC buffer. For GenBank accession numbers 30 and primer sequences refer to Supplemental Table 1. PCR products were sequenced by LGC 31 Genomics (Berlin, Germany) or on an ABI 3730xl Genetic Analyzer (Life Technologies). 32 Base-pair substitutions were verified twice, frameshift mutations once. Multiple frameshifts in 33 a single PCR product were analyzed in detail by subcloning into Topo TA cloning vectors 34 (Life Technologies) and subsequent sequencing. 35 Statistical Analysis 36 Probability of event-free survival (pEFS) was calculated according to Kaplan and Meier with 37 differences compared by log-rank test. pEFS was calculated from date of diagnosis to first 38 event (death from any cause, relapse, resistant disease or second malignancy) or to the date of 39 last follow-up. Patients lost to follow-up were censored at date of last follow-up-examination. 40 Cumulative incidence functions for relapse were constructed by the method of Kalbfleisch 41 and Prentice, and compared with Gray’s test. Differences in the distribution of individual 42 parameters among patient subsets were analyzed using χ² or Fisher’s exact test. Supplementary Figures Supplementary Figure 1: Probability of EFS according to PI3K-AKT pathway mutational status. Supplementary Figure 2: Probability of EFS according to NOTCH1 mutation (a) and loss of heterozygosity at chromosome 6q (LOH6q) status (b). Supplementary Tables Supplementary Table 1: Primer sequences used for amplification of PIK3R1, PIK3CA, PTEN, NRAS and KRAS. Some of the primers have been published.4-8 Region PIK3R1 exons 16+17 PIK3CA exon 10 exon 21 PTEN exon 7 KRAS exon 2 NRAS exon 2 exon 3 Forward primer Reverse primer Product length (bp) GCTGGGAAACCATAGTGAAA CTACAAGATGTTCCAAACTCAG 548 TTGCTTTTTCTGTAAATCATCTGTG TGGGGTAAAGGGAATCAAAAG CTGCTTTATTTATTCCAATAGGTATG CCTATGCAATCGGTCTTTGC 321 525 TGACAGTTTGACAGTTAAAGG CTTCTCAGTTAACCATCCTTG 466 AAGCGTCGATGGAGGAGTTT TGGACCCTGACATACTCCCA 545 GGCCGATATTAATCCGGTGT AACAGCACAAATAAAACAGTCCAG TCCGACAAGTGAGAGACAGG TGGTAACCTCATTTCCCCATA 332 627 Supplementary Table 2: Detailed description of identified mutations. Numbering of base pairs and amino acids according to GenBank accession indicated. patient T-LBL-5 T-LBL-7 T-LBL-11 PIK3R1 NM_181523.2, NP_852664.1 DNA protein PIK3CA NM_006218.2, NP_006209.2 DNA protein T-LBL-18 T-LBL-21 c.1781G>A T-LBL-25 T-LBL-28 T-LBL-41 c.3297A>G T-LBL-44 T-LBL-46 c.3302G>C T-LBL-48 PTEN NM_000314.4, NP_000305.3 DNA protein c.1808C>A p.H259Q c.[1752_1781 delins CCCGCGGA A]+ [1727_1728in sG] p.R233fsAT GRQVHVL* p.F241_C250 delinsPAE c.[1727_1728 insT]+ [1732_1733in sGCCG]+ [1732_1733in sATAGTAG G] p.R233fsSTG RQVHVL* p.E235fsPGR QVHVL* p.E235* c.1728_1730 delins GCCCCCCG GGG p.R233fsAPR GGKTSSCT LSSLSRYLC VVISK* homozygous mutation c.[1768_1769 insTCCACC] + [1727_1728in s TTGTAAT] p.P246_L247 insPP p.R231fsL* KRAS NM_004985.4, NP_004976.2 DNA protein c.38G>A p.Gly13Asp c.35G>C p.Gly12Ala NRAS NM_002524.4, NP_002515.1 DNA protein p.E542K p.H1047R p.G1049R c.35G>A p.Gly12Asp patient T-LBL-49 T-LBL-52 PIK3R1 NM_181523.2, NP_852664.1 DNA protein PIK3CA NM_006218.2, NP_006209.2 DNA protein c.2315_2320 del CAATAC p.Q579_Y58 0del c.1781G>A p.E542K c.2270A>G p.N564D c.3144A>G p.N996S c.2270A>G p.N564D c.1781G>A p.E542K T-LBL-53 T-LBL-55 T-LBL-57 T-LBL-58 T-LBL-60 T-LBL-62 c.1728_1729i nsA T-LBL-65 c.1768delins GGGGCCCA T-LBL-74 T-LBL-80 T-LBL-82 T-LBL-85 c.2326-2A>G T-LBL-87 c.2270A>G T-LBL-92 PTEN NM_000314.4, NP_000305.3 DNA protein c.[1728delins p.R234fsDG AGG]+ KTSSCTLSS [1762_1763in LSRYLCVVI sTCAGCCG SK* ACGG p.L247fsTGR GAAGACA QVHVL* AGTTCATG p.P246fsRRY TACTT LCVVISK* TGAGTTCC p.P246_L247 C]+ insGWR [1767_1768in sGG]+ [1769_1770in sGGTTGGA GG] c.1753_1754 p.F241* delTT c.1727_1728i p.R233fsTTG nsA RQVHVL* c.1729delins p.R233fsGT GG GRQVHVL* c.3297A>G KRAS NM_004985.4, NP_004976.2 DNA protein NRAS NM_002524.4, NP_002515.1 DNA protein c.34G>A p.Gly12Ser c.34G>A p.Gly12Ser c.38G>T p.Gly13Val c.35G>A p.Gly12Asp p.R233fsTTG RQVHVL* homozygous mutation p.P246fsRGP VTCVW* p.H1047R p.M582_D60 5delinsI p.N564D patient T-LBL-94 T-LBL-95 T-LBL-100 T-LBL-103 T-LBL-105 T-LBL-111 T-LBL-112 T-LBL-113 T-LBL-114 PIK3R1 NM_181523.2, NP_852664.1 DNA protein PIK3CA NM_006218.2, NP_006209.2 DNA protein PTEN NM_000314.4, NP_000305.3 DNA protein c.[1726_1728 delinsTGGG] + [1767_1768in sAA] c.1768_1769i nsCC p.P246fsQR YLCVVISK* p.T232fsMG TGRQVHVL * p.L247fsRYL CVVISK* c.[1728delins GA]+ [1768delinsG GGGG] c.[1729_1749 delinsCCG]+ [1732G>C] p.R233fsETG RQVHVL* p.P246fsRG VTCVW* p.R233_Y24 0delinsPD p.R234P c.1725_1727 delinsCCAG C p.T232fsPAD GKTSSCTLS SLSRYLCV VISK* p.R233fsAT GRQVHVL* c.1727_1728i nsG KRAS NM_004985.4, NP_004976.2 DNA protein c.35G>T p.Gly12Val NRAS NM_002524.4, NP_002515.1 DNA protein c.181C>A p.Gln61Lys c.183A>T p.Gln61His c.34G>T p.Gly12Cys Supplementary Table 3: Frequency and outcome according to the mutational status of PI3K, PTEN, RAS, NOTCH1, FBXW7 and loss of heterozygosity of chromosome 6q (LOH6q). PI3K, combined data for PIK3R1, PIK3CA; RAS, combined data for KRAS and NRAS; Data on mutations of NOTCH1, FBXW7 and LOH6q have been published earlier. Marker PI3KWT PI3Kmut PTENWT PTENmut RASWT RASmut NOTCH1WT NOTCH1mut FBXW7WT FBXW7mut LOH6qneg LOH6qpos n 98 9 96 17 89 10 46 70 95 21 192 25 EFS (5 years) 77±4% 78±14% 82±4% 59±12% 78±4% 70±14% 66±7% 84±5% 76±5% 79±10% 86±3% 27±9% p value (log rank) 0.99 0.014 0.61 0.021 0.6 <0.0001 Supplementary Table 4: Comparison of published classifier for adult T-ALL9 with data on adult T-ALL (a), applied to our cohort of pediatric T-LBL (b) and our newly defined genetic classifier for pediatric T-LBL (c). CIR, cumulative incidence of relapse; CI, confidence interval. a) Definition NOTCHmut ± FBXW7mut + RASWT + PTENWT all other patients N evaluable patients 91 81 CIR (5 years) 95% CI p 15% 54% 9–24% 42–66% <0.001 CIR (5 years) 95% CI p 16% 23% 6–27% 11–35% 0.28 CIR (5 years) 95% CI p 11% 19% 64% 1–22% 7–31% 37–91% <0.001 b) Definition NOTCHmut ± FBXW7mut + RASWT + PTENWT all other patients N evaluable patients 51 48 c) Definition GR: NOTCH1mut + RASWT + PI3KWT IR: all other patients HR: NOTCH1WT + PTENmut ± LOH6qpos N evaluable patients 35 42 14 Supplementary References 1. Oschlies I, Burkhardt B, Chassagne-Clement C, d’Amore ES, Hansson U, Hebeda K, et al. Diagnosis and immunophenotype of 188 pediatric lymphoblastic lymphomas treated within a randomized prospective trial: experiences and preliminary recommendations from the European childhood lymphoma pathology panel. Am J Surg Pathol 2011; 35: 836-844. 2. Reiter A, Schrappe M, Ludwig WD, Tiemann M, Parwaresch R, Zimmermann M, et al. Intensive ALL-type therapy without local radiotherapy provides a 90% event-free survival for children with T-cell lymphoblastic lymphoma: a BFM group report. Blood 2000; 95: 416-421. 3. Bonn BR, Rohde M, Zimmermann M, Krieger D, Oschlies I, Niggli F, et al. Incidence and prognostic relevance of genetic variations in T-cell lymphoblastic lymphoma in childhood and adolescence. Blood 2013; 121: 3153–3160. 4. Chang YS, Yeh KT, Hsu NC, Lin SH, Chang TJ, Chang JG. Detection of N-, H-, and KRAS codons 12, 13, and 61 mutations with universal RAS primer multiplex PCR and N-, H-, and KRAS-specific primer extension. Clin Biochem 2010; 43: 296-301. 5. Li VS, Wong CW, Chan TL, Chan AS, Zhao W, Chu KM, et al. Mutations of PIK3CA in gastric adenocarcinoma. BMC cancer 2005; 5: 29. 6. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004; 304: 554. 7. Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 2010; 42: 338-342. 8. Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest 2008; 118: 3762-3774. 9. Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengliné E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult Tcell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol 2013; 31: 4333–4342.