Fig. S1. DNA sequence analysis of the myomaker promoter in
advertisement

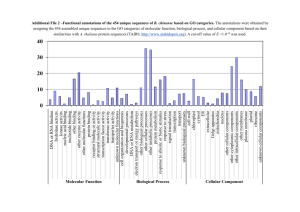
1 Fig. S1. DNA sequence analysis of the myomaker promoter in multiple animals to find putative NFAT binding sites. The sequence 1500 bp upstream of the (a) squirrel and mouse, and (b) squirrel and human myomaker transcriptional start sites were interrogated using rVista for the detection of NFATc1-4 binding sequences. Red lines indicate aligned hits of possible NFATc1-4 binding regions between organisms, the two sequences highlighted were identified by rVista as possible binding sites. Cross-referencing with NFAT binding sequences reported in the literature (c, shown in green), one probable binding site remained 1095 bp upstream of the myomaker transcriptional start site. This sequences along with its aligned flanking region from the multiple alignments was used to design a 25 bp probe for DNA ELISA. Fig. S2. (a) Amino acid multiple alignment using ClustalW2 for myomaker proteins from human, mouse, and squirrel showing strong sequence conservation. * indicates identical residues, : indicates conservation between amino acids of similar properties and . represents dissimilar properties. Red letters represent hydrophobic amino acids, blue – acidic, magenta – basic, green – hydroxyl, sulfhydryl, or amine. (b) Pair-wise amino acid alignment percent identities were determined using ClustalW2 for squirrel, mouse, and human NFATc1-4, myoferlin, and myomaker proteins.