longterm_results
advertisement

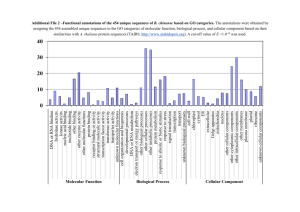
Aadel Chaudhuri Long Term Project Results Determination of Let-7 Target Sites The let-7 search on the invertebrate 3' UTR database yielded sixty-seven predicted targets out of a total of 26,664 UTR fasta entries. Of these, 31 have known developmental roles, while 36 have other or unknown roles. Of the targets with developmental roles, six are known to be heterochronic, three are involved in neuronal remodelling, five have vulval functions, and five are nuclear hormone receptors. The functions of the top 20 targets are all developmental, with the exception of one which has unknown function (figure 2) [14]. The binding interaction between let-7 and its two highest-ranked target UTR sites (Daf-12 and hunchback) is shown in figure. Thermodynamic analysis of let-7 binding with all possible target sites reveals values of free energy of binding ranging from -85 kJ/mol to -20 kJ/mol (figure 3) [12-13]. In comparison, free energy of binding of the ten randomized sequences was on average 15 kJ/mol higher (figure 4). This indicates that the let-7 miRNA would bind to its targets more strongly than the randomized sequences would bind to their targets. Analyses of lin-4 Binding Interactions Thermodynamics of binding between lin-4 and its target sequences revealed values between -105 kJ/mol and -18 kJ/mol (figure 5). The curve correlating free energy and UTR entry is quite a bit steeper for lin-4 than for any of the random sequences. Still, the 40 strongest target sites have free energy values of binding that are on average 35 kJ/mol lower than those of the scrambled sequences. The top lin-4 target site was the transcript of the lin-14 gene, a heterochronic gene with developmental roles. Lin-4 and let-7 binding sites on the lin-14 UTR are shown in figure 6 [11-12].