Lo-Ciganic et al, Machine Learning and Medication Adherence
advertisement

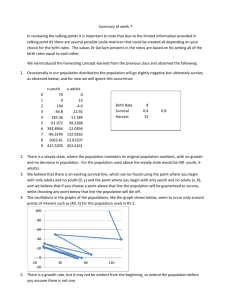
Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement Online Supplement eMethod. Technical Appendix eTable 1. Operational Definitions for Diabetes-related Hospitalizations eTable 2. Hospitalization Rates during the Post-Index Year eTable 3. PDC and Hazard Ratios for Each Terminal Node in Survival Tree: AllCause Hospitalizations eTable 4. Multivariate Cox Proportional Models with Same Set of Predictors in Survival Tree: All-Cause Hospitalizations eTable 5. PDC and Hazard Ratios for Each Terminal Node in Survival Tree: Diabetes-Related Hospitalizations eTable 6. Multivariate Cox Proportional Models with Same Set of Predictors in Survival Tree: Diabetes-Related Hospitalizations eFigure 1. Sample Size Flow Chart eFigure 2. Important Predictors of Diabetes-related Hospitalizations Selected by Minimal Depth from Random Survival Forests eFigure 3. Adherence Thresholds associated with Risk of Diabetes-related Hospitalizations: A Survival Tree 1 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eMethod. Technical Appendix Random Survival Forests Details of random survival forest techniques are described elsewhere and in the technical appendix.1-3 To select the most important predictors of hospitalizations, we constructed a random survival forest of 1,000 survival trees, where each tree was from an independent and unique bootstrap sample of the training sample. At each branch or node, a random set of candidate predictors were chosen as candidates to split the node into 2 other branches, and the number of variables assessed at each branch was the square root of the total number of variables (e.g., the square root of 14, which was rounded as 4.). For each of the randomly selected variables, the variable whose split yielded the highest log-rank value was chosen to occupy the first node.4 The categorical variables were split according to their categories and continuous variables were split at randomly selected cut points (nsplit=5 in our study). For each subsequent node of the tree, random selection of candidate predictors and selection of the best split or threshold were repeated. The process continued until we reached a unique subset that contained no fewer than three hospitalization events (Figure A).2 From these individual survival trees, we identified important variables by averaged minimal depth from the tree trunk for each variable.2 The most predictive variables were defined as those whose average minimal depth (i.e., split nodes nearest to the root node) is smaller than the minimal depth of a variable which was unrelated to the survival distribution and determined under the null hypothesis of no effect (threshold).5 The smaller the minimal depth, the greater the association with the dependent variable and hence the impact of that variable on prediction. Simply due to random chance, a variable with no prediction power may on occasion split at a lower depth when a large number of trees are grown. Thus, similar to prior work, we used the average minimal depth of such an unrelated variable as a threshold to identify a set of important predictors whose average minimal depths were less than the said threshold. These important predictors were further used to construct a survival tree described in the next section. We assessed the prediction accuracy of random survival forests by the Harrell concordance index (C-index) using the out-of-bag method (i.e., bootstrap 2/3 sample of the training sample).2,6 C-index is defined as the probability of concordance given that the pairs considered are usable in which at least one had an event. It can be interpreted as the probability that a patient from the event group has a higher predicted probability of having an event than a patient from the non-event group. Unlike other measures of survival performance, Harrell’s C-index does not depend on choosing a fixed time for evaluation of the model and specifically takes into account censoring of individuals.7 A small prediction error is preferred; however, there is no gold standard how small would be desirable. Previous studies have reported prediction error rates ranged from 25-40%, which indicates some promise for the predictors, but not ideal and indicative of the complexity of the predicting health outcomes. In these studies, random survival trees performed at least as good as or better than traditional models.8,9 Survival Trees We then fit survival trees with the important predictors identified from random survival forests and explored the optimal threshold of adherence to oral hypoglycemics that was most strongly associated with hospitalizations.10 Briefly, survival trees start with the root that included all patients from the training sample and used binary recursive partitioning methods to systematically search among all predictors for variables that classify or segment a target population into increasingly homogeneous subgroups with respect to the outcome of interests (i.e., hospitalizations in this study). For continuous variables (e.g., PDC), it searched the threshold value that optimally split patients into groups with similar likelihood of hospitalization risk. Partitioning stopped when risks for hospitalizations for the two partitioned subgroups were not statistically different based on log-rank tests or the minimum terminal node size was less than 20 patients. In addition, we used 10-fold cross-validation methods to guard against model over-fitting (complex parameter=0.005). We calculated hazard ratios (HR) for each terminal node. To compare prediction performance, we compared C-indices with 95% confidence intervals (CIs) between the final survival tree and Cox proportional hazard model.6,11 2 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement 3 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eMethod. Technical Appendix (Continued) Figure A. Illustration of a Random Tree from Random Survival Forests. A bootstrap sample of patients from the original dataset is used to build a random tree. At the open circles randomly selected subset of variables (e.g., PDC, age) compete to split node. Among these, single variable that discriminates between event/non-event best chosen to permanently split node. Node levels are numbered based on their relative distance to the root of the tree (i.e., level 1, 2, 3). Splitting of nodes to create the tree continues until terminal nodes have few distinct events. Each terminal node (*) contains a group of patients with unique characteristics, and a survival curve demonstrating their outcome. 4 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 1. Operational Definitions for Diabetes-related Hospitalizations Type of diabetes-related hospitalizations Diabetes Hyperglycemia Hypoglycemia Septicemia or bacteremia Pneumonia Kidney infections, cystitis, urinary tract infection Cellulitis Electrolyte imbalance Diabetes retinopathy ICD-9 or CPT codes 250.xx 250.1x, 250.2x, 250.3x 250.8x, 251.1x, 251.2x 038.xx, 790.7 480-6 590, 595, 599.0x 680-682, 686 276.xx 250.5x, 361.xx, 362.0x, 362.1, 362.8x, 379.23, 369.xx 250.4x, 585.xx, 593.9 250.6x, 356.9, 357.2x 410-414, v45.81, v45.82; CPT codes: 36.1x, 36.2x, 00.66, 36.06, 36.07 433-434 250.7x, 440.2x, 707.1x, 785.4x, v49.6, v49.7; CPT codes: 84.0x, 84.1x [excluded if any diagnosis is 895-897] Diabetic nephropathy Diabetic neuropathy Ischemic heart disease Stroke Diabetes peripheral circulatory disorders 5 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 2. Hospitalization Rates during the Post-Index Year Hospitalizations Training sample (N=29,855) All-cause hospitalizations, n (%) 1 ≥2 Number of months to first all-cause hospitalizations, mean (SD)/median (minmax) Diabetes-related hospitalizations, n (%)a 1 ≥2 Number of months to first diabetes-related hospitalizations, mean (SD)/median (minmax) Testing sample (N=3,275) 4,224 (14.2) 478 (14.6) 2,936 (9.8) 331 (10.1) 5.2 (3.5)/ 4.7 (0.03-12) 5.2 (3.6)/ 4.9 (0.03-12) 2,822 (9.4) 327 (10.0) 1,124 (3.8) 118 (3.6) 5.6 (3.5)/ 5.6 (0.03-12) 5.7 (3.6)/ 5.3 (0.03-12) a : Diabetes-related hospitalizations were defined by inpatient admission during the post-index year with an ICD-9 codes as “primary” discharge diagnosis or current procedural terminology codes in any position, including diabetes, hyperglycemia, hypoglycemia, septicemia or bacteremia, pneumonia, kidney infections, cystitis, urinary tract infection, cellulitis, electrolyte imbalance, diabetes retinopathy, diabetic nephropathy, diabetic neuropathy, ischemic heart disease, stroke, and diabetes peripheral circulatory disorders 6 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 3. PDC and Hazard Ratios for Each Terminal Node in Survival Tree: AllCause Hospitalizations Terminal nodes A B C D E F G H I J K L M N O P Q N 7,989 1,186 3,367 956 303 2,130 1,728 481 291 4,198 484 1,462 1,879 554 1,347 694 806 Average PDC (SD) 0.63 (0.26) 0.66 (0.25) 0.76 (0.22) 0.87 (0.10) 0.45 (0.13) 0.82 (0.12) 0.38 (0.13) 0.78 (0.21) 0.97 (0.01) 0.50 (0.25) 0.92 (0.04) 0.47 (0.21) 0.78 (0.16) 0.32 (0.10) 0.82 (0.13) 0.37 (0.13) 0.72 (0.23) 7 Medium PDC (min, max) HR (95% CI) 0.66 (0.03, 1.00) 0.68 (0.03,1.00) 0.84 (0.03, 1.00) 0.91 (0.63, 1.00) 0.47 (0.14, 0.62) 0.84 (0.60, 1.00) 0.40 (0.04, 0.59) 0.85 (0.16, 100) 0.96 (0.95, 1.00) 0.49 (0.03, 0.94) 0.92 (0.84, 1.00) 0.48 (0.04, 0.83) 0.81 (0.47, 1.00) 0.33 (0.07, 0.46) 0.85 (0.57, 1.00) 0.39 (0.03, 0.56) 0.79 (0.07, 1.00) Referent 1.42 (1.22, 1.64) 1.41 (1.28, 1.56) 1.78 (1.54, 2.06) 3.16 (2.59, 3.84) 1.64 (1.46, 1.83) 2.40 (2.16, 2.67) 3.12 (2.67, 3.69) 0.94 (0.67, 1.30) 1.94 (1.78, 2.12) 1.88 (1.54, 2.28) 3.03 (2.74, 3.38) 2.71 (2.46, 3.00) 3.93 (3.44, 4.56) 3.43 (3.10, 3.83) 5.54 (4.98, 6.30) 6.02 (5.46, 6.79) Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 4. Multivariate Cox Proportional Models with Same Set of Predictors in Survival Tree: All-Cause Hospitalizations Prior hospitalizations or ED visits Had insulin fills during the index year (reference= non-users) 0-<90 days ≥ 90 days Had diabetes comorbidities (ref=DCSI=0) PDC Number of monthly total prescriptions C-statistics (error rate)* HR (95% CI) 1.67 (1.59, 1.76) P value <0.0001 1.41 (1.29, 1.55) 1.27 (1.20, 1.34) 1.42 (1.35, 1.49) <0.0001 <0.0001 <0.0001 0.53 (0.48, 0.58) 1.06 (1.06, 1.07) <0.0001 <0.0001 0.672 (32.8%) Abbreviations: DCSI: diabetes comorbidity severity index; ED: emergency department; HR: hazard ratios; PDC: proportion of days covered * Error rate for the survival tree was 26% 8 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 5. PDC and Hazard Ratios for Each Terminal Node in Survival Tree: DiabetesRelated Hospitalizations Terminal nodes A B C D E F G H I J K L M N O N Average PDC (SD) Medium PDC (min, max) HR (95% CI) 11,356 2,869 426 1,889 1,695 793 1,304 990 2,440 1,612 1,181 612 683 1,069 936 0.67 (0.26) 0.52 (0.27) 0.96 (0.02) 0.59 (0.23) 0.82 (0.12) 0.81 (0.12) 0.35 (0.14) 0.39 (0.13) 0.84 (0.11) 0.39 (0.14) 0.77 (0.21) 0.89 (0.08) 0.47 (0.17) 0.82 (0.12) 0.37 (0.14) 0.72 (0.03, 1.00) 0.51 (0.03, 1.00) 0.96 (0.93, 1.00) 0.62 (0.03, 0.92) 0.84 (0.60, 1.00) 0.81 (0.60, 1.00) 0.34 (0.07, 0.59) 0.41 (0.04, 0.59) 0.87 (0.62, 1.00) 0.41 (0.04, 0.59) 0.84 (0.11, 1.00) 0.91 (0.73, 1.00) 0.49 (0.09, 0.72) 0.84 (0.60, 1.00) 0.37 (0.03, 0.59) Referent 1.31 (1.14, 1.51) 1.25 (0.89, 1.73) 2.27 (1.98, 2.60) 1.75 (1.51, 2.05) 2.72 (2.28, 3.27) 2.52 (2.18, 2.95) 3.68 (3.19, 4.27) 1.74 (1.51, 2.00) 2.71 (2.39, 3.14) 4.04 (3.56, 4.65) 2.62 (2.14, 3.21) 4.02 (3.51, 4.87) 4.79 (4.20, 5.47) 6.64 (5.94, 7.64) 9 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eTable 6. Multivariate Cox Proportional Models with Same Set of Predictors in Survival Tree: All-Cause Hospitalizations Prior hospitalizations or ED visits Had insulin fills during the index year (reference= non-users) 0-<90 days ≥ 90 days Had diabetes comorbidities (ref=DCSI=0) PDC Number of monthly total prescriptions C-statistics (error rate)* HR (95% CI) 1.52 (1.42, 1.62) P value <0.0001 1.72 (1.53, 1.93) 1.71 (1.60, 1.83) 1.65 (1.54, 1.77) <0.0001 <0.0001 <0.0001 0.52 (0.46, 0.58) 1.07 (1.05, 1.06) <0.0001 <0.0001 0.669 (33.1%) Abbreviations: DCSI: diabetes comorbidity severity index; ED: emergency department; HR: hazard ratios; PDC: proportion of days covered * Error rate from the survival tree was 29% 10 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement Total beneficiaries in the Pennsylvania Medicaid Program during 2007-2011 (N=3,720,189) Of these, patients who were non-dual eligible to Medicare and Pennsylvania residents during 2007-2011 (N=3,007,749) Of these, patients enrolled at least for 30 months consecutively during 2007-2011 (study period), and were at the age of 18-64 years at the index year (N= 454,992) Exclude those who were (1) dual eligible to Medicare (n=616,244), and then (2) not Pennsylvania residents (n=96,196) Exclude those who were (1) not consecutively enrolled for at least 30 months during 2007-2011 (n=1,819,233), and then (2) aged under 18 or 65 and older at the beginning of enrollment (n=733,524) Exclude non-diabetic patients (n=366,566) Patients had a diagnosis for diabetes (ICD9= 250.xx) or had any prescription fills for diabetes medications during study period (N=88,426) Of these, patients had a diagnosis for T2DM (ICD-9 codes= 250.0x-250.9x, where x=0 or 2) and had any prescription fills for OHA during study period (N=47,201) Of these, patients had an index date of first OHA fill between 07/01/2007-12/31/2009 and at least 2 prescription fills for non-insulin anti-diabetic medications during index year (N=33,994) Of these, patients who did not have nursing home stay ≥ 90 days during the index year (N=33,130) Exclude patients who were (1) with T1DM (n=1,042), (2) with gestational diabetes (n=1,974), (3) without any diabetes diagnosis and only had insulin fills (n=767), (4) had same numbers of claims with T1DM and T2DM diagnoses, and only with insulin fill (n=54), (5) had more than half of the claims with diagnoses of T1DM and only insulin fills, [n=706]), (6) without any anti-diabetic medication fills (n=31,526), and then (7) T2DM but only with insulin (n= 5,076), or pramlintide/exenatide (n=80) fills during 2007-2011 Exclude patients who (1) did not have any OHA fills after 07/01/2007 (n=1,025), (2) had an index date after 12/31/2009 (n=9,643), (3) did not have any baseline period before index date (n=14), (4) had only one prescription fill for OHA medications during the index year (n=2,405), (5) age >65 at the index date (n=47), and then (6) died during the index year (n=73) Exclude patients who (1) were long-term institutionalized (≥ 90 days nursing home stay during the index year (n=444), and then (2) were hospitalized ≥ 90 days during the index year (n=420) eFigure 1. Sample Size Flow Chart Abbreviations: OHA: oral hyperglycemic agents; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus Note: We included three other exclusion criteria but were not listed in the chart because n=0: (1) women who used metformin only, had a diagnosis for polycystic ovary syndrome, but no diagnosis for diabetes, (2) hyperglycemia not otherwise specified (ICD-9 790.6 without any diabetes code). 11 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement eFigure 2. Important Predictors of Diabetes-related Hospitalizations Selected by Minimal Depth from Random Survival Forests Note: From 1,000 individual survival trees, the most predictive variables were defined as those whose average minimal depth (i.e., split nodes nearest to the root node) is smaller than the minimal depth of a variable which was unrelated to the survival distribution and determined under the null hypothesis of no effect (i.e., threshold). The threshold was calculated from a variable whose distribution of average minimal depth behaves a random coin tossing experiment, or average minimal depth increase little while number of variables increases. The horizontal dashed line in the figure is the threshold for filtering variables. All variables below the line are important predictors. 12 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement Error rate: 29% eFigure 3. Adherence Thresholds associated with Risk of Diabetes-related Hospitalizations: A Survival Tree Abbreviations: DCSI: diabetes complication severity index; ED: emergency department; HR=hazard ratio; PDC: proportion days covered Note: Nodes (A) to (O) are terminal nodes. The numbers on the bottom of each node represent numbers of events/patients in that node. PDCs were median PDC of patients in each terminal node. 13 Lo-Ciganic et al, Machine Learning and Medication Adherence Thresholds: Online Supplement REFERENCES 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. Ishwaran H, Kogalur UB. Package ‘randomForestSRC’. 2014; http://cran.rproject.org/web/packages/randomForestSRC/index.html. Accessed October 30, 2014. Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2(3):841-860. Ishwaran H, Kogalur UB. Random survival forests for R. Rnews. 2007;7:25-31. Segal MR. Regression trees for censored-data. Biometrics. 1988;44:35-47. Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-Dimensional Variable Selection for Survival Data. J Am Stat Assoc. 2010;105(489):205-217. Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361-387. May M, Royston P, Egger M, Justice AC, Sterne JA, Collaboration ARTC. Development and validation of a prognostic model for survival time data: application to prognosis of HIV positive patients treated with antiretroviral therapy. Stat Medicine. 2004;23(15):2375-2398. Taylor JM. Random Survival Forests. J Thorac Oncol. 2011;6(12):1974-1975. Chen G, Kim S, Taylor JM, et al. Development and validation of a quantitative real-time polymerase chain reaction classifier for lung cancer prognosis. J Thorac Oncol. 2011;6(9):14811487. Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression trees. 1 ed. Belmont, CA: Wadsworth International Group; 1984. Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109-2123. 14