bit25244-sm-0001-SuppData-S1
advertisement
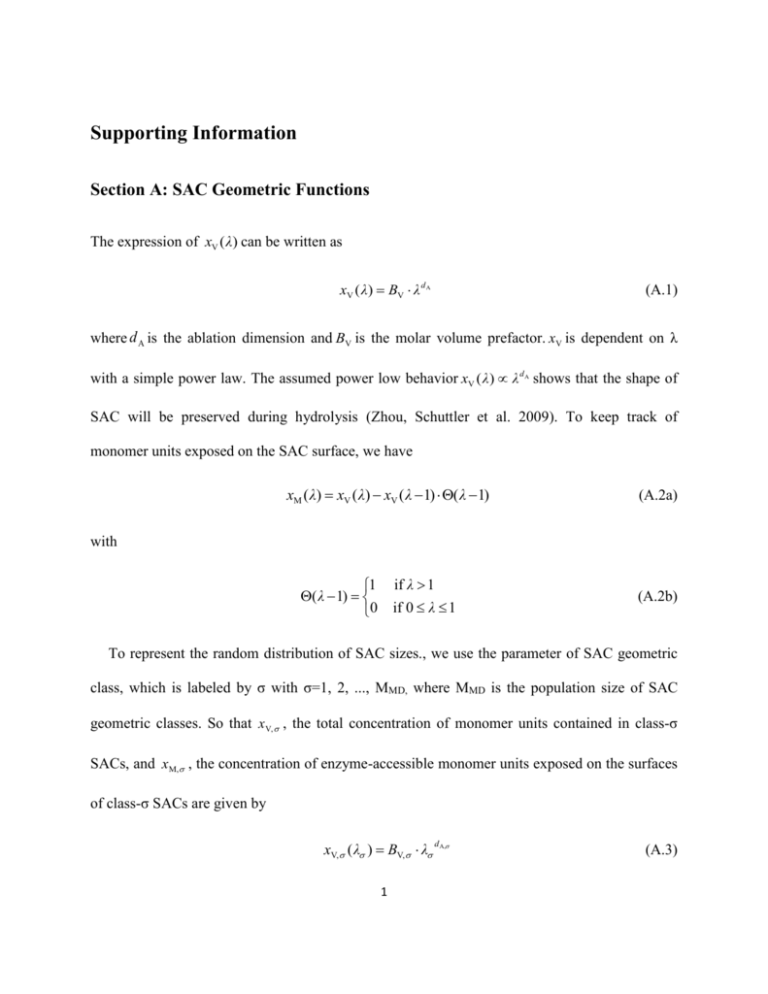
Supporting Information Section A: SAC Geometric Functions The expression of xV ( λ) can be written as xV ( λ) BV λ d A (A.1) where d A is the ablation dimension and BV is the molar volume prefactor. xV is dependent on λ with a simple power law. The assumed power low behavior xV ( λ) λ dA shows that the shape of SAC will be preserved during hydrolysis (Zhou, Schuttler et al. 2009). To keep track of monomer units exposed on the SAC surface, we have xM ( λ) xV ( λ) xV ( λ 1) ( λ 1) (A.2a) with if λ 1 if 0 λ 1 1 ( λ 1) 0 (A.2b) To represent the random distribution of SAC sizes., we use the parameter of SAC geometric class, which is labeled by σ with σ=1, 2, ..., MMD, where MMD is the population size of SAC geometric classes. So that x V, , the total concentration of monomer units contained in class-σ SACs, and xM, , the concentration of enzyme-accessible monomer units exposed on the surfaces of class-σ SACs are given by xV, ( λ ) BV, λ 1 d A, (A.3) xM, ( λ ) xV, ( λ ) xV, ( λ 1) ( λ 1) (A.4a) with 1 ( λ 1) 0 if λ 1 if 0 λ 1 (A.4b) where BV, is the molar volume prefactor for class-σ SACs, and d A, is the ablation dimension factor for class-σ SACs. x V, and xM, must obey the relationships written as xV xV, ( λ ) (A.5) xM xM, ( λ ) (A.6) We use two parameters to describe the enzyme accessibility, which are FA , the overall fraction of enzyme-accessible monomer units, and FA, , the fraction of enzyme-accessible monomer units for the class-σ SACs. Their expressions are given by FA, ( λ ) FA xM, ( λ ) xV, ( λ ) xM FA, ( λ ) xV (A.7) (A.8) where xV, / xV . The value of FA can be directly observed before enzymatic hydrolysis and (0) used as an initial input value, that is, FA , in the model, and decide the initial value of . We use the assumption of the Gaussian distribution for the total molar concentration of monomer units per geometric class, and then have 2 Avg wid (0) exp ( Avg 2 )2 / 2 / exp ( ) / 2 wid (A.9) By adjusting the two Gaussian distribution parameters Avg and Wid , a proper initial distribution of the molar monomer concentration per geometry class can be obtained to match the value of FA(0) . Section B: Derivation of Surface Layer Ablation Rate Equations In order to derive the surface layer ablation rate equations, we first define R , , 0 as the loss rate of type-μ sites belonging to type-ρ chains exposed on class-σ SAC surfaces, and R 0 as the total loss rate of monomer units exposed on class-σ SAC surfaces into solution. So that we can obtain R R , , , d xV, ( λ (t )) λ xV, ( λ ) λ dt (B.1) where λ is shorthand for the derivative / λ . The removal of a small fraction of a layer during ) a short time interval dt will result in a removal of dxV,( fra λ xV, dλ monomer units from the SAC surface by chain fragmentation and then result in a change of SAC exposed surface area ( fra ) by dxM, λ xM, dλ monomer units. The net concentration of newly exposed monomer units at the surface, resulting from the removal of overlaying monomer units and the change of surface area, is thus (exp) ( fra ) ( fra ) dxM, dxV, dxM, ( λ xV, λ xM, )dλ R (1 λ xM, / λ xV, )dt Based on Eq. (B.2), the net concentration of newly exposed sites is given by 3 (B.2) (exp) dx(exp) , , (dxM, ) M, , ( 1) g , , (B.3) where g , , ( ) is the native site fraction functions representing the fraction of type-μ sites contained in type-ρ chains in the interior of the class-σ SACs. g , , ( ) can be treated as a layer–independent parameter, which means g , , ( ) g , , ( ) g , , . In addition, we denote g E , , as the native L-end (or R-end) broken site fraction of type-ρ chains in class-σ SACs, which has such relationship g i LUi , , i g RU , , g E , , and can be obtained from the i degree of polymerization of type-ρ chains DP . Combining the surface exposure contribution dx(exp) , , with the surface fragmentation ) contribution dx( fra , , , which is given by ) dx( fra , , R , , dt (B.4) ) (exp) we can obtain the net increment of type-μ surface sites, that is, dx , , dx( fra , , dx , , , and the rate function Equation (3a). Then from Eq. (B.1) we can immediately obtain the changing rate of , which is Equation (3b). 4 Section C: Enzyme Adsorption and Inhibition Equilibriums The first step for an enzyme molecule to cut an intact bond is to form an enzyme–substrate (ES) complex, that is, to adsorb a bond site exposed on SAC surfaces. The cutting rate for a intact bond site of type μ in a chain belonging type ρ, having already adsorbed an enzyme molecule on the surface of an SAC belonging class σ, is thus given by the product of rate coefficient and ES complex concentration, written in the form , μ , z κ , μ , ,σ , where κ represents the type of enzymes; z κ , μ , ,σ represents the concentration of (κ , μ, ) ES complexes on class-σ SAC surfaces; and , μ , represents the cutting rate coefficient (in the unit of cuts per second per ES complex), which is identical for all the geometric classes. It is assumed that the ES complex formation process is much faster than the bond cutting kinetics. Therefore, the enzyme adsorption quasi-equilibrium is maintained at the SAC surfaces during hydrolysis and written as z κ , μ , ,σ Lκ , μ , vκ y μ , ,σ (C.1a) where y μ , ,σ is the molar concentration of free type-μ sites contained in type-ρ chains on class-σ SAC surfaces, vκ is the concentration of free type-κ enzymes and Lκ , μ , is the substrate adsorption coefficient which is the inverse of the conventional desorption equilibrium coefficient. As hydrolysis proceeds, some enzyme molecules may also be inhibited by soluble oligomers to form enzyme-oligomer (EO) complexes in solution. This type of enzyme adsorption equilibrium in solution can be written as zκ , (l ) I κ, (l ) vκ yS , (l ) 5 (C.2b) with 1 l lS , . Here, zκ , (l ) is the concentration of (κ , , l ) EO complexes in which each oligomer contains l monomer units, I κ , (l ) is the oligomer adsorption coefficient; yS , (l ) is the molar concentration of free oligomers contained l monomer units and dissolved from type-ρ chains, and vκ is the same parameter as in Equation (C.1a). As described in Supporting Information section B, zκ , μ , ,σ and zκ , (l ) can be calculated and expressed in terms of x , , and xS , (l ) by solving enzyme adsorption and inhibition equations. The construction of RS , (l ) describes the reactions between oligomers and beta-enzymes and requires the solutions of the enzyme adsorption and inhibition equilibriums. In this model, the reaction mechanism of beta-enzymes in solution is simplified as: a free oligomer which contains l monomer units can adsorb and be hydrolyzed by its corresponding beta-enzymes into a monomer unit and an oligomer containing l 1 monomer unit(s). So the expression of RS , (l ) can be written as lS , 1 R (1) (2) z (2) S , 2 , (l ) zκ , (l ) , κ , RS , (l ) , (l ) zκ , (l ) , (l 1) zκ , (l 1) RS , (lS , 1) , (lS , 1) zκ , (lS , 1) l lS , 1 2 l lS , 2 l lS , 1 (C.3) where , (l ) is the oligomer cutting rate coefficient. Note that in this model, only beta-enzymes could hydrolyze oligomers in solution after forming EO complexes with them in solution. So for endo- and exo- enzymes, their corresponding values of , (l ) are 0, which means they are inhibited by forming EO complexes with oligomers. And for any monomer unit, , (1) is also 0 6 which means all the enzymes will inevitably be inhibited by monomer units during hydrolysis. Furthermore, both the cutting rate coefficient , (l ) and the oligomer adsorption coefficient I κ , (l ) have two dimensions representing the enzyme types and the chain types. The free enzyme and surface site concentrations vκ and y μ , ,σ are related to their total concentrations u κ and x μ , ,σ , respectively, by way of the total enzyme and total site balance relations: uκ vκ z μ , ,σ κ , μ , ,σ zκ , (l ) (C.4) ,l xS , (l ) yS , (l ) zκ , (l ) (C.5) x μ , ,σ y μ , ,σ zκ , μ , ,σ κ κ , μ ', ' f μ , ,σ κ zκ , μ ', ',σ (C.6) where f μ, ,σ xμ, ,σ / xM ,σ . The last term in the Equation (C.6) arises from the fact that the footprint area of a surface-adsorbed enzyme molecule is far greater than the average surface area of a bond site. Hence, a type-κ enzyme molecule, bound to a type-μ' surface site, will in effect cover up, and obstruct access to some number ( ) of other surface sites that are located in spatial proximity to the type-μ' binding site. Due to the current lack of value of for each type enzymes discussed in this work, we simply use the value of cellulase, that is, 39, for all the enzymes. Equations. (C.1a), (C.2b), (C.4), (C.5) and (C.6) can then be solved simply by iteration in order for vκ , y μ , ,σ , z κ , μ , ,σ and zκ , (l ) . Combining the mass action and balance relations, the general form of enzyme adsorption and inhibition equilibrium can be shown as 7 vκ 1 μ , ,σ uκ Lκ , μ , yμ , ,σ I κ , (l ) yS , (l ) (C.7) ,l yS , (l ) xS , (l ) 1 I κ , (l ) vκ (C.8) y μ , ,σ xμ , ,σ μ , (1 μ ', ' μ ', ', μ ', ' ) μ , 1 L , μ, v (C.9) (C.10) μ , L , μ , v (C.11) μ , , xμ , , μ , xM , (C.12) To calculate vκ and y μ , ,σ , as functions of u κ and x μ , ,σ this system of coupled non-linear equations can be solved iteratively, starting from the initial guess v (init ) u ( init ) yS , (l ) xS , (l ) (init ) y ,μ , x , μ , (C.13) Section D: Site Number Increments and Chain Site Distribution Model N ', , (k , k ' ) can be expressed as N , , (k , k ' ) N , , (k ) N , , (k ' ) N , , (k k ' ) 8 (D.1) where N , , (k ) denotes the average number of type-μ sites per insoluble type-ρ chain of class-σ SACs for k lS , , with N , , (k ) 0 for k lS , ,and N , , (k , k ' ) is thus the difference of the number of type-μ sites belonging to type-ρ chain caused by a cut. The specific functional form of N , , (k ) depends on the distribution of site types along a type-ρ chain. So we construct the site distribution model based on the assumption that O-sites are randomly distributed with a uniform probability φO , ,σ over all l 1 intact bonds within a type-ρ chain. By straightforward site counting, we thus construct N , , (l ) for any insoluble type-ρ chain with l lS , 7 (l 1) φ O , ,σ φUi , N , , (l ) (1 φO , ,σ ) φUi , φU j , (l 3) (1 φO , ,σ ) φUi , φU j , l φ Ji , μO μ LUi or RUi μ X Uij or YUij (D.2) μ NUij μ Ji Here, φU i , is the average molar ratio of the type-i monomer units to all the monomer units contained in the backbone of a type-ρ chain, and φJ i , is the average molar ratio of the type- J i side groups to all the monomer units contained in the backbone of a type-ρ chain. Both of them actually characterize the natural composition of the type-ρ chains which can be easily obtained from literature and used in the model. Based on Equations (D.1) and (D.2),we can easily obtain the detailed expression of N ', , (k , k ' ) . 9 Section E: Derivation of Chain Fragmentation Probabilities This section shows the derivation of how to express P , (k , k ' | , 1) in terms of P , (l ) and P , ( μ | k , k ' ,1) . Here, P , (l ) is defined as the probability of a randomly selected insoluble type-ρ chain, exposed on a class-σ SAC surface, to contain l monomer units, and P , ( μ | k , k ' ,1) is the probability for a randomly selected intact bond to be of site type μ on a type-ρ chain, provided that the site is located k monomer units from the L-end and k' monomer units from the R-end of the type-ρ chain. P , ( μ | k , k ' ,1) describes the distribution of different site types along a type-ρ chain (relative to the chain ends). In order to express P , (k , k ' | , 1) in terms of P , (l ) and P , ( μ | k , k ' ,1) , first, let us consider a random sample of type-ρ chains, with random chain lengths and a very large sample size NL →∞. Let these NL type-ρ chains be concatenated, in random order, into a "super type-ρ chain" where the R-end of each individual chain is connected to the L-end of its right neighbor chain by a fictitious bond, referred to as a “−1-bond”, and the real internal bonds between monomer units contained in each chain are referred to as “+1-bonds”. Hence, we are assigning to each bond site on the “super type-ρ chain” a “bond integrity” variable ζ, with ζ = +1 for intact bond site types NUij , X Uij , YUij , O and ζ = −1 for broken bond site types between adjacent chain ends (i.e. for a pair of adjacent L,R-sites). Then, let us consider the average chain length for type-ρ main chains l , , that is, the average degree of polymerization (DP), for type-ρ chains exposed on class-σ SAC surfaces. 10 l , can be expressed in terms of the L-end broken site fraction of type-ρ chains f E , , , or the concentration of type-ρ chain xE , , , which is written as l , l P , (l ) 1/ f E, , xM, , / xE, , (E.1) l 1 In Equation (E.1), f E , , comes from the site type fractions f , , , which is, for any site type belonging to type-ρ chains on class-σ SAC surfaces, defined by f , , x , , (E.2) xM , , Based on the "uniform segment exposure" assumption (Zhou, Schuttler et al. 2009), the number of left chain ends must equal the number of right chain ends, which is x LUi , , i xRU , , xE , , . Also, because of the relationship between LU i and XUij site groups, i i and between RU i and YUij site groups, we can get f i f i RUi , , fYU f E , , i, j x i ij , , i, j ij / (1 φO , ,σ ) and , , / (1 φO , ,σ ) , so that f E , , can be expressed as LUi , , xM , , f XU LUi , , f LU , , i i f i, j X Uij , , (1 φO , ,σ ) f i, j YUij , , (1 φO , ,σ ) f RU , , i i x i RUi , , xM , , (E.3) Next, we denote P , (k , k ' , ) as the probability that a bond randomly selected from the "super type-ρ chain" is a ζ-bond, where ζ is either +1 or −1, and that this randomly selected ζ-bond will be located k ≥ 1 monomer units from its nearest L-end and be located k' ≥ 1 monomer units from its nearest R-end. The expression of P , (k , k ' , ) is given by: 11 P , (k , k ', 1) f E , , P , (k k ') P , (k , k ', 1) f E , , P , (k ) P , (k ') (E.4) This can be derived from the "super type-ρ chain" construction as follows: for ζ = +1, P , (k , k ' ,1) is a joint probability for two conditions meeting at the same time: the first one is that the bond located k monomer units to the left of the randomly selected ζ-bond should be a “−1-bond”; the second one is that the adjacent k + k' monomer units to the right of that “−1bond” should form a single contiguous chain of length l k k ' . The probabilities for these two conditions are f E , , and P , (k k ' ) , respectively, hence the joint probability P , (k , k ' ,1) is the product f E , , P , (k k ') . For ζ = −1, P , (k , k ' ,1) can be derived by a similar way. And it can be easily verified that k ,k '1 1 P , (k , k ', ) 1 . Next, we introduce the conditional site type probability, given the type-ρ chain fragments, denoted by P , ( | k , k ' , ) which is the probability for a randomly selected "super type-ρ chain" bond to be of site type μ, given that the randomly selected bond is a ζ-bond; and given that it is located k and k' monomer units from its nearest −1-bond to the left and to the right, respectively. Just like N , , (k ) , the conditional type-ρ chain site probabilities P , ( | k , k ' , ) depend on the distribution of type-μ sites along the type-ρ chains. In fact, for purposes of the fragmentation kinetics P , ( | k , k ' , ) comprises the complete mathematical description of the chain site distribution model. The values of P , ( | k , k ' ,1) and N , , (k ) are not independent of each other: for any type-ρ chain site distribution, P , ( | k , k ' , ) must be normalized to P ( | k , k ', ) 1 , and P ( | k , k ' ,1) and N (k ) must obey the following general chain , , , , site number counting relations for all intact bond site types: 12 l 1 N , , (l ) (l lS , 1) P , ( | k , l k ,1) (E.5a) k 1 for NUij , X Uij , YUij , O with 1 if l 0 (l ) 0 if l 0 (E.5b) where P , ( | k , k ' ,1) completely determines N , , (k ) for site groups NUij , X Uij , YUij , O . Based on the chain site model N , , (l ) , we thus assign the site groups NUij , X Uij , YUij , O of the intact type-ρ chain bonds to the corresponding "super type-ρ chain" +1-bonds, while formally LU i , RU i is randomly assigned to each "super type-ρ chain" −1-bond with probability φU i , ,σ / 2 corresponding to 1 / 2 for site groups LU and RU . Then P , ( | k , k ' , ) can be written as i i φ ,1 O , ,σ ,1 φUi , ,σ / 2 P , ( μ | k , k ', ) ,1 k ,k X (1 φO , ,σ ) φUi , ,σ φU j , ,σ ,1 k ',kY (1 φO , ,σ ) φUi , ,σ φU j , ,σ (1 k , k X , ) (1 k ', kY , ) (1 φO , , σ ) φU i , , σ φU j , , σ ,1 μO μ LUi or RUi μ X Uij (E.6) μ YUij μ NUij Finally, we can construct the conditional type-ρ fragmentation probability, given the site type, P , (k , k ' | , ) , defined as the probability for a randomly selected bond in “super type-ρ chain” to be located k and k' monomer units from its nearest “−1-bond” to the left and to the right, respectively, given that the bond is a ζ-bond and that it is of site type μ. By Bayes’ theorem, we have P , (k , k ' | , ) P , ( | k , k ', ) P , (k , k ', ) P , ( , ) 13 (E.7) where the unconditional site type probability P , ( , ) is given by: P , ( , ) ,1 f , , P ( k , k ', , ) , k , k '1 ,1 f , , / 2 for =NUij , X Uij , YUij , O for =LUi , RUi (E.8) P , (k , k ' | , ) is defined only where P , ( , ) 0 . The fragmentation probabilities are normalized as k 1 k '1 P , (k , k ' | , ) 1 .Thus we obtain the expression of P , (k , k ' | , 1) in terms of P , (l ) and P , ( μ | k , k ' ,1) , given by P , (k , k ' | , 1) f E , , f , , P , ( | k , k ', 1) P , ( k k ') (E.9) By using Equation (E.9), the expression of NS , , , (k ) can be written as ( N S , , , (k ) ( ( f E , , f , , f E , , f , , f E , , f , , ) Ui , U j , (1 O , , ) 2 k ,k X , k ,kY , P , (k k X , ) P , (k kY , ) NU ij ) Ui , U j , (1 O , , ) k ,k X , P , ( k k X , ) XU ) Ui , U j , (1 O , , ) k ,kY , P , (k kY , ) YU ij ij (E.10) Section F: Detailed Expression of the Surface Sites Increment Factor N ', , , 14 ( ( ( ( ( ( ( ( ( ( ( ( ( ( ( f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , f E , , f , , ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 P , (lS , ) P , (lS , k X , -1) XU , ' XU ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 P , (lS , ) P , (lS , k X , -1) X U , ' YU ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 P , (lS , ) P , (lS , kY , -1) YU , ' X U ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 P , (lS , ) P , (lS , kY , -1) YU , ' YU 1 ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 2lS , -1 f E , , 1 ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 2lS , -1 f E , , ij ij i' j' ij ij i' j' i' j' i' j' NU , ' X U NU , ' YU ij ij i' j' lS , k X , -1 ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 k X , (l - k X , - 3) P , (l ) l lS , X U , ' NU lS , kY , -1 ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 kY , (l - kY , - 3) P , (l ) l lS , YU , ' NU lS , k X , -1 lS , kY , -1 3 ) -U i , U j , U i ' , U j ' , (1 O , , ) 2 lS , 2 - 7lS , 3 - k X , - kY , ( k X , 3 - l ) P , (l ) ( kY , 3 - l ) P , (l ) f l lS , l lS , E , , NU , ' NU lS , k X , -1 ) -U i , U j , (1 O , , ) O , , k X , (l - k X , -1) P (l ) l lS , XU , ' O lS , kY , -1 ) -U i , U j , (1 O , , ) O , , kY , (l - kY , -1) P (l ) l lS , YU , ' O ij ij i' j' i' j' ij ij ij lS , k X , -1 lS , kY , -1 1 ) -U i , U j , (1 O , , ) O , , ls 2 - 3ls 1- k X , - kY , ( k X , 1- l ) P (l ) ( kY , 1- l ) P (l ) f l ls l ls E , , lS , k X , -1 ) -U i , U j , (1 O , , ) Ji , , k X , (l - k X , ) P (l ) l ls NU , ' O ij XU , ' Ji ij lS , kY , -1 ) -U i , U j , (1 O , , ) J i , , kY , (l - kY , ) P (l ) l ls YU , ' J i lS , k X , -1 lS , kY , -1 ) -U i , U j , (1 O , , ) J i , , lS , 2 - lS , - k X , - kY , ( k X , - l ) P (l ) ( kY , - l ) P (l ) l ls l ls NU , ' J i ij 15 ij (F.1) i' j' i' j' Section G: Derivation of Production Factor of Oligomer As P , (k , k ' | , 1) can be expressed in terms of P , (l ) and P , ( μ | k , k ' ,1) , we can obtain N S , , , (k ) ( f E , , f , , ) k ' ls , k P , ( | k , k ', 1) P , ( | k ', k , 1) P , (k ' k ) (G.1) By using the "Chain End Decomposition" theory (Zhou, Schuttler et al. 2009) we can separate the effects of the near-chain-end sites, which can be cut by both exo- and endo-enzymes, from the chain interior sites, which can only be cut by endo-enzymes. P , ( | k , k ', 1) can then be decomposed into left end (L), right end (R) and interior (I) contributions of the type-ρ main chain, which can be written as P , ( | k , k ', 1) P(,I) , L, (k ) P(,L), (k ) R, (k ') P(,R), (k ') (G.2) where L , (k ) (lL , k ) and R , (k ') (lR , k ') are the cut-off factors dividing a type-ρ chain into three parts with lL , k X , 1 and lR , kY , 1 . Notice that in the model, k k ' lLR , lL , lR , 1 and P , ( | k , k ', 1) P(,I) , is independent of k and k ' when there are k lL , and k ' lR , . Based on the functional form, we can obtain in detail the values of three contributions for each hydrolysable intact bond site, shown as P(,I) , (1 φO , ,σ )φU , φU , i j ( L ) P , , (k ) k ,k (1 φO , ,σ )φU , φU , X , i j ( R ) P , , (k ') k ',k (1 φO , ,σ )φU , φU , Y , i j (I ) ( L) P , , 0; P , , (k ) k ,k X , (1 φO , ,σ )φUi , φU j , ; P(,R), (k ') 0 P(,I) , 0; P(,L), (k ) 0; P(,R), (k ') k ',kY , (1 φO , ,σ )φU i , φU j , 16 μ NUij μ X Uij μ YU ij (G.3) which then can be used to obtain the detailed expressions of NS , , , (k ) . Section H: Derivations of Chain Concentration Rate Equations and Rate Equations Closure In order to find the solution for P , (l ) with l lC , , a new rate equation system is developed for type-ρ chains concentration variables H , (l ) by defining H , (l ) P , (l ) xE , , , with H , (l ) 0 for l lS , . Here, xE , , is the concentration of insoluble type-ρ chains, as discussed in Supporting Information section D. All the surface site concentration variables now could be xM , , l l l H , (l ) expressed in terms of H , (l ) by using S , and x , , l l N , , (l ) H , (l ) . Analogous to the site ablation rate equations, the rate equations S , of H , (l ) is given by H , (l ) RH , , (l ) R ( ) M , , ( 1) Q , (l , 1) / j lS , j Q , ( j, 1) (H.1) where RH , , (l ) is the changing rate of type-ρ chains of length l due to bonds cutting, and the second term of the equation above gives the rate of exposure of new type-ρ chains due to the removal of overlaying material. Q , (l , λ ) is the type-ρ chain length distribution of the native substrate material in each layer of class-σ SACs, which obeys the delta-function native type-ρ chain length distribution Q , (l , λ ) l , DP . Q , (l , λ ) has a close relationship with the native site fraction g , , , which is given by 17 g E , , 1/ l l Q , (l , λ ) S , g , , ( ) g E , , l N , , (l ) Q , (l , λ ) (H.2a) S , (H.2b) As described in Supporting Information section G, RH , , ( k ) can be written by RH , , (l ) , , z , , , N H , , , (k ) , (H.3) Then we can have RH , , (l ) R , (l k , k ') R , ( j k , l ) R , ( j l , k ) k , k '1 k 1 j l 1 (H.4) and R , (l k , k ') , , z , , , P , (k , k ' | , 1) l , k k ' , (H.5) R , (l k , k ') is denoted as the rate at which surface-exposed type-ρ chains of length l on class-σ SACs are being cut into two type-ρ chain fragments of length k and k ' , from the L-end and R-end of the original type-ρ chains respectively. As P , (k , k ' | , 1) can be expressed in terms of P , (l ) and P , ( μ | k , k ' ,1) , we can rewrite RH , , ( k ) by RH , , (l ) , , z , , , N H , , , (k ) , (H.6) By using the "Chain End Decomposition" (Zhou, Schuttler et al. 2009) theory P , ( | k , k ', 1) , we can obtain 18 N H , , , (l ) ( f E , , f , , ) AH , , , (l ) BH , , , (l ) DH , , , (l ) (H.7a) where AH , , , (l ) N , , (l ) P , (l ) l BH , , , (l ) 2P(,I) , P(,L), (l )L, (l ) P(,R), (l )R, (l ) 1 j 1 P , ( j ) DH , , , (l ) l lE , P , (l ) P(,L), ( j l ) L, ( j l ) P(,R), ( j l ) R , ( j l ) j l 1 lE , max(lL , , lR , ) 1 (H.7b) (H.7c) (H.7d) The detailed expression of N H , , , (l ) can be obtained by combining Equations (H.7a), (H.7b), (H.7c), (H.7d) and (D.2), written as ( N H , , , (l ) ( ( f E , , f , , f E , , f , , f E , , f , , l ) Ui , U j , (1 O , , ) 2 2 P , ( k ) (l 3) P , (l ) P , (l k X , ) P , (l kY , ) k 1 NU ij ) Ui , U j , (1 O , , ) P , (l k X , ) P , (l ) XU ) Ui , U j , (1 O , , ) P , (l kY , ) P , (l ) YU ij (H.8) To solve the chain concentration rate equations for H , (l ) at short chain lengths from l to the cut-off length lC , , it is necessary to use the value of P , ( j ) at chain lengths from l to lC , lE , . Obviously, the length ranges of l are different for H , (l ) and P , ( j ) , which will make the equation system redundant. So in order to solve H , (l ) with the length range of lS , l lC , by using P , (l ) with the same length range, the Local Poisson (LP) approximation scheme (Zhou, Schuttler et al. 2009) is used here for P , (l ) , so that we have 19 ij P , (l ) P , (lC , ) P , (lC , ) / P , (lC , 1) ( l lC , ) (H.9) for lC , 1 l lC , lE , . By using Equation (H.9) with the relationship P , (l ) H , (l ) / xE , , , a closed ordinary differential equation (ODE) system for the site concentration formalism is completed. Section I: Parameters All parameters are categorized into four groups and showed in four tables respectively. The (0) (0) first group in Table I1 includes FA and M,Xyl , which describe the initial substrate morphology i.e. the enzymatic accessibilities of the whole substrate and xylan, respectively. Based on the values of these two parameters we can obtain the enzymatic accessibility of cellulose. We believe that the values of these two parameters could be measured by using reliable experimental techniques in the future. However, there are currently no values for these two parameters. So we (0) adjust the values for FA and M,Xyl based on the values of enzymatic accessibility of cellulose, (0) which is commonly measured in experiments. The values we used were from Zhu et al. (2009). Based on the experiments by Zhu et al. (2009) the enzymatic accessibility of cellulose was 0.243 for DA-pretreated corn stover and 0.0238 for non-pretreated corn stover. Thus we believe that the value of the enzymatic accessibility of cellulose for AFEX-pretreated corn stover, as well as poplar, should range from 0.0238 to 0.243. The values of the enzymatic accessibility of cellulose we used in the model were 0.0832, 0.0998 and 0.0753. The second group is adsorption and kinetic parameters and shown in Table I2. The values of L1, N ,Glu L1, X ,Glu L1,Y ,Glu L2, N ,Glu L2,Y ,Glu L2, X ,Glu L3, N ,Glu L3, X ,Glu L , , , , , , , and 3,Y ,Glu , which describe the 20 adsorption equilibrium between cellulose-sites and cellulases, were adopted from Zhou et al. (2009) with referenced experiments. and L3,Y , Xyl L1, N , Xyl L1, X , Xyl L1,Y , Xyl L2, N , Xyl L2, X , Xyl L2,Y , Xyl L3, N , Xyl L3, X , Xyl , , , , , , , are parameters describing the adsorption equilibrium between xylan sites and cellulases. The values for these parameters were all set to be 0 as we assume that if there is no effective adsorption between enzyme molecule and bond site leading to bond cleavage, the parameter of adsorption equilibrium would be set to 0. For the same reason, L4, N ,Glu L4, X ,Glu , , L4,Y ,Glu L5, N ,Glu L5, X ,Glu L5,Y ,Glu L5, N , Xyl L5,Y , Xyl L7, , L , , , , , , and 8, , , which describe the adsorption equilibrium between sites and enzymes having no adsorption relationships, were also set to be 0. L4, N , Xyl L4, X , Xyl L , and 4,Y , Xyl describe the adsorption equilibrium between Endo-acting xylanases and xylan sites and the values of these parameters were from the experiments by Qing and Wyman (2011). The values of kinetic parameters 4,Y , Xyl 1, N ,Glu 1, X ,Glu 1,Y ,Glu 2, X ,Glu 3,Y ,Glu 4, N , Xyl 4, X , Xyl , , , , , , and came from the experiments by Banerjee et al. (2010) which used AFEX-pretreated corn stover as substrate. The relationship among these parameters was based on Zhang and Lynd (2006) in which the activity ratio of EG:CBH2:CBH1 was 5:2:1. For ineffective adsorptions that cannot lead to bond cleavage, the corresponding kinetic parameters would be 0. So the values of 1, N , Xyl 1, X , Xyl 1,Y , Xyl 2, N ,Glu 2,Y ,Glu 2, N , Xyl 2, X , Xyl 2,Y , Xyl 3, N ,Glu 3, X ,Glu 3, N , Xyl 3, X , Xyl 3,Y , Xyl , , , , , , , , , , , , , 4, N ,Glu 4, X ,Glu , and 4,Y ,Glu were all set to be 0. Currently, there are no reliable values of the adsorption and kinetic parameters of Exo-acting xylanases. So we assumed that values of L5, N ,Glu , L5, X ,Glu L5,Y ,Glu L5, N , Xyl L5,Y , Xyl L5, X , Xyl L6, N ,Glu L6, X ,Glu L6,Y ,Glu L6, N , Xyl L6, X , Xyl L , , , , , , , , , and 6,Y , Xyl were equal to the values of corresponding parameters of Endo-acting xylanases. However, these parameters 21 did not affect the simulation results since it is assumed that commercial enzyme mixtures do not contain significant amount of Exo-acting xylanases. The third group is inhibition parameters and shown in Table I3. I (1) I1,Glu (1) I 2,Glu (1) , and 3,Glu are D-glucose (G1) inhibition parameters for EG, CBH2 and CBH1 respectively, and their values were from Levine, Fox et al. (2010). Similarly, I1,Glu (2) I 2,Glu (2) I (2) , and 3,Glu are cellobiose (G2) inhibition parameters for EG, CBH2 and CBH1 respectively, and their values were from Jeffrey et al. (1999). There are limited reported values for cello-oligomers (G3-G6) inhibition parameters for cellulases. The values of I1,Glu (3) I 2,Glu (3) I 3,Glu (3) I1,Glu (4) I 2,Glu (4) I 3,Glu (4) , , , , , , I (6) I (2) I1,Glu (5) I 2,Glu (5) I 3,Glu (5) I1,Glu (6) I 2,Glu (6) , , , , and 3,Glu were based on the values of 1,Glu , I 2,Glu (2) and I 3,Glu (2) as well as the experiments by Lo Leggio and Pickersgill (1999) which describe the relationship among cello-oligomers (G2-G6) inhibition parameters for cellulases. Currently, there are no reliable values of the D-xylose (X1) inhibition parameters for EG, CBH2 and CBH1 respectively. So we assumed that values of I (1) I1, Xyl (1) I 2, Xyl (1) , and 3, Xyl were equal to I (1) I1,Glu (1) I 2,Glu (1) , and 3,Glu respectively due to the similarity between the structures of these two monomer units. I (2) I1, Xyl (2) I 2, Xyl (2) , and 3, Xyl are xylobiose (X2) inhibition parameters for EG, CBH2 and CBH1 respectively, and their values were from the work by Ntarima et al. (2000). The values of I1, Xyl (3) I 2, Xyl (3) I 3, Xyl (3) I1, Xyl (4) I 2, Xyl (4) I 3, Xyl (4) I1, Xyl (5) I 2, Xyl (5) I 3, Xyl (5) , , , , , , , , , I (6) I (2) I1, Xyl (6) I 2, Xyl (6) I (2) I (2) , and 3, Xyl were based on the values of 1, Xyl , 2, Xyl and 3, Xyl as well as the experiments by Lo Leggio and Pickersgill (1999) which also describe the relationship among xylo-oligomers (X2-X6) inhibition parameters for cellulases. 22 I 4,Glu (1) is D-glucose (G1) inhibition parameter for Endo-acting xylanases which currently do not have too much reliable values. So we assumed that value of I 4,Glu (1) was equal to I1,Glu (1) due to the same inhibitor. For the same reason, the values of cello-oligomers (G2-G6) inhibition parameters for Endo-acting xylanases, which are I 4,Glu (2) I 4,Glu (3) I 4,Glu (4) I 4,Glu (5) I (6) , , , and 4,Glu , were all determined. The values of D-xylose (X1) and xylobiose (X2) inhibition parameters for Endo-acting xylanases I 4, Xyl (1) and I 4, Xyl (2) were based on the work by Ntarima et al. (2000). The values of I 4, Xyl (3) , I (6) I 4, Xyl (4) I 4, Xyl (5) I (2) , and 4, Xyl were based on the value of 4, Xyl and the experiments by Lo Leggio and Pickersgill (1999) which described the relationship among xylo-oligomers (X2-X6) inhibition parameters for Endo-acting xylanases. Currently, there are no reliable values for the inhibition parameters of Exo-acting xylanases. So we assumed that the values of inhibition parameters of Exo-acting xylanases were equal to the values of corresponding inhibition parameters of Endo-acting xylanases. However, these parameters did not affect the simulation results since we did not consider Exo-acting xylanases as enzyme species in any commercial enzyme. The fourth group is parameters about beta-enzymes and shown in Table I4. I 7,Glu (2) I 7,Glu (1) and are the D-glucose (G1) adsorption (or inhibition) parameter for beta-glucosidase (BG) and the cellobiose (G2) adsorption parameter for BG respectively. The values of these parameters were from the experiment by Chauve et al. (2010). I 7,Glu (6) I 7,Glu (3) , I 7,Glu (4) , I 7,Glu (5) and are cello-oligomers (G3-G6) adsorption parameters for BG. Their values were based on the value of I 7,Glu (2) and the experiments by Yazaki et al. (1997) which described the 23 relationship among cello-oligomers (G2-G6) inhibition parameters for BG. I 8, Xyl (1) I 8, Xyl (2) , , I (6) I 8, Xyl (3) I 8, Xyl (4) I 8, Xyl (5) , , and 8, Xyl are D-xylose and xylo-oligomers (X2-X6) adsorption parameters for beta-xylosidase (BX). Their values were from the work by Rasmussen, Sorensen et al. (2006). Currently, there are no reliable values for the xylo-oligomers (X1-X6) adsorption parameters for BG and cello-oligomers (G1-G6) inhibition parameters for BX. We only considered the inhibitions of X1 for BG and G1 for BX and did not consider other "crossover" oligomers adsorption for beta-enzymes in the model. We assumed that the values of I 8,Glu (1) were equal to BG kinetic parameter I 8, Xyl (1) 7,Glu (2) and I 7,Glu (1) I 7, Xyl (1) and respectively due to the same inhibitors. The value of for cellobiose (G2) was from experiment by Chauve et al. (2010). 7,Glu (3) 7,Glu (4) 7,Glu (5) (6) , , and 7,Glu are BG kinetic parameters for cello-oligomers (G3-G6), their values were based on the value of 7,Glu (2) and the experiments by Yazaki et al. (1997) which described the relationship among BG kinetic parameters for cello-oligomers (G2-G6). 8, Xyl (2) 8, Xyl (3) 8, Xyl (4) 8, Xyl (5) (6) , , , and 8, Xyl are BX kinetic parameters for xylo-oligomers (X2-X6) and their values from the work by Rasmussen et al. (2006). As we did not consider other "crossover" oligomers adsorption for beta-enzymes in the model, the corresponding kinetic parameters were all set to be 0. 24 Table.I1 Key simulation parameters Qing and Wyman 2011 Banerjee, Car et al. 2010 Kumar and Wyman 2009 Parameter Value Parameter Value Parameter Value FA(0) 0.1051 FA(0) 0.1431 FA(0) 0.1046 (0) M,Xyl 0.55 (0) M,Xyl 0.50 (0) M,Xyl 0.60 Table.I2 Adsorption and kinetic parameters Parameter Value Ref. Parameter (1/mM) L1, N ,Glu L1, X ,Glu L1,Y ,Glu 3 Value Ref. (1/min) 1, N ,Glu 1, X ,Glu 1,Y ,Glu (Zhou, 3317 (Zhang and Hao et Lynd 2006, al. 2009) Banerjee, Car et al. 2010) L1, N , Xyl L1, X , Xyl L1,Y , Xyl 1, N , Xyl 1, X , Xyl 1,Y , Xyl 0 0 (Zhou, Hao et al. 2009) L2, N ,Glu L2,Y ,Glu 0 2, N ,Glu 2,Y ,Glu 0 L2, X ,Glu 4 2, X ,Glu 1399 (Zhang and Lynd 2006, Banerjee, Car et al. 2010) 25 L2, N , Xyl L2, X , Xyl L2,Y , Xyl 2, N , Xyl 2, X , Xyl 2,Y , Xyl 0 0 (Zhou, Hao et al. 2009) L3, N ,Glu L3, X ,Glu 0 3, N ,Glu 3, X ,Glu 0 L3,Y ,Glu 3 3,Y ,Glu 699.7 (Zhang and Lynd 2006, Banerjee, Car et al. 2010) L3, N , Xyl L3, X , Xyl L3,Y , Xyl 3, N , Xyl 3, X , Xyl 3,Y , Xyl 0 0 (Zhou, Hao et al. 2009) L4, N ,Glu L4, X ,Glu L4,Y ,Glu 0 L4, N , Xyl L4, X , Xyl L4,Y , Xyl 0.574 (Zhang 4, N ,Glu 4, X ,Glu 4,Y ,Glu 0 4, N , Xyl 4, X , Xyl 4,Y , Xyl 50.12 (Zhang and and Lynd 2006, Lynd Banerjee, 2006, Car et al. Qing and 2010) Wyman 2011) 5, N ,Glu 5,Y ,Glu 5, X ,Glu 0 5, N , Xyl 5,Y , Xyl 0 0.574 5, X , Xyl 50.12 L6, N ,Glu L6, X ,Glu L6,Y ,Glu 0 6, N ,Glu 6,Y ,Glu 6, X ,Glu 0 L6, N , Xyl L6, X , Xyl 0 6, N , Xyl 6, X , Xyl 0 L6,Y , Xyl 0.574 6,Y , Xyl 50.12 L7, , L8, , 0 7, , 8, , 0 L5, N ,Glu L5, X ,Glu L5,Y ,Glu 0 L5, N , Xyl L5,Y , Xyl 0 L5, X , Xyl Assu med 26 Assum ed Table.I3 Inhibition parameters Parameter Value Reference Parameter (1/mM) I1,Glu (1) 0.06 Value (1/mM) (Levine, Fox I 2,Glu (1) I 3,Glu (1) 0.032 et al. 2010) I1,Glu (2) 0.13 (Jeffrey Tolan 0.3 S. I 2,Glu (2) I 3,Glu (2) 0.13 and (Jeffrey Tolan I1,Glu (4) 0.37 I1,Glu (5) 0.44 S. Leggio 0.51 S. and Foody 1999) I 2,Glu (3) I 3,Glu (3) 0.3 I 2,Glu (4) I 3,Glu (4) 0.37 I 2,Glu (5) I 3,Glu (5) 0.44 and I1,Glu (6) (Jeffrey Tolan and Foody 1999, Lo (Levine, Fox et al. 2010) Foody 1999) I1,Glu (3) Reference (Jeffrey S. Tolan and Foody 1999, Lo Leggio and Pickersgill I 2,Glu (6) I 3,Glu (6) 0.51 Assumed I 2, Xyl (1) I 3, Xyl (1) 0.032 (Lo I 2, Xyl (2) I 3, Xyl (2) 2 Pickersgill 1999) 1999) I1, Xyl (1) 0.06 I1, Xyl (2) 2 Leggio and I1, Xyl (3) 2 I1, Xyl (4) 4 10 Pickersgill 1999, I1, Xyl (6) 11 I 4,Glu (1) 0.06 Nerinckx (Lo Leggio and Pickersgill I 2, Xyl (3) I 3, Xyl (3) 2 I 2, Xyl (4) I 3, Xyl (4) 4 Ntarima, I1, Xyl (5) Assumed 1999, Ntarima, Nerinckx et al. 2000) et I 2, Xyl (5) I 3, Xyl (5) 10 I 2, Xyl (6) I 3, Xyl (6) 11 I 5,Glu (1) I 6,Glu (1) 0.032 al. 2000) Assumed 27 Assumed I 4,Glu (2) 0.13 (Jeffrey Tolan I 4,Glu (3) 0.3 I 4,Glu (4) 0.37 S. 0.13 I 5,Glu (3) I 6,Glu (3) 0.3 I 5,Glu (4) I 6,Glu (4) 0.37 I 5,Glu (5) I 6,Glu (5) 0.44 I 5,Glu (6) I 6,Glu (6) 0.51 I 5, Xyl (1) I 6, Xyl (1) 0.4 I 5, Xyl (2) I 6, Xyl (2) 0.85 I 5, Xyl (3) I 6, Xyl (3) 1.5 I 5, Xyl (4) I 6, Xyl (4) 2 I 5, Xyl (5) I 6, Xyl (5) 4 I 5, Xyl (6) I 6, Xyl (6) 4.5 and Foody 1999, Lo I 5,Glu (2) I 6,Glu (2) Leggio and I 4,Glu (5) 0.44 I 4,Glu (6) 0.51 I 4, Xyl (1) 0.4 Pickersgill 1999) (Ntarima, Nerinckx et al. 2000) I 4, Xyl (2) 0.85 I 4, Xyl (3) 1.5 (Lo Leggio and Pickersgill I 4, Xyl (4) 2 1999, Ntarima, I 4, Xyl (5) 4 I 4, Xyl (6) 4.5 Nerinckx et al. 2000) Table.I4 Beta-enzymes parameters Parameter Value Reference Parameter (1/mM) I 7,Glu (1) 0.294 Value Reference (1/min) (Chauve, 7,Glu (1) 0 7,Glu (2) 1897 Determined Mathis et al. 2010) I 7,Glu (2) 1.136 (Yazaki, Ohnishi et al. (Chauve, Mathis et al. 28 1997, I 7,Glu (3) 3.846 2010) 7,Glu (3) Chauve, 1738.9 Mathis et al. I 7,Glu (4) 4.000 I 7,Glu (5) 2.174 2010) (Yazaki, Ohnishi et al. 7,Glu (4) 1422.8 7,Glu (5) 895.8 1997, Chauve, Mathis et al. 2010) I 7,Glu (6) 1.449 I 7, Xyl (1) 0.417 I 7, Xyl (2) 0 7,Glu (6) 843.1 Assumed 7, Xyl (1) 0 Not 7, Xyl (2) 0 Determined considered I 7, Xyl (3) 0 7, Xyl (3) 0 I 7, Xyl (4) 0 7, Xyl (4) 0 I 7, Xyl (5) 0 7, Xyl (5) 0 I 7, Xyl (6) 0 7, Xyl (6) 0 I 8,Glu (1) 0.294 Assumed 8,Glu (1) 0 I 8,Glu (2) 0 Not 8,Glu (2) 0 considered I 8,Glu (3) 0 8,Glu (3) 0 I 8,Glu (4) 0 8,Glu (4) 0 I 8,Glu (5) 0 8,Glu (5) 0 I 8,Glu (6) 0 8,Glu (6) 0 I 8, Xyl (1) 0.417 8, Xyl (1) 0 8, Xyl (2) 1897 (Rasmussen, Sorensen I 8, Xyl (2) 2.500 al. 2006) et 29 (Rasmusse I 8, Xyl (3) 5.000 8, Xyl (3) 1250.3 n, Sorensen et al. 2006) I 8, Xyl (4) 6.250 8, Xyl (4) 1164.1 I 8, Xyl (5) 10.000 8, Xyl (5) 1293.4 I 8, Xyl (6) 12.500 8, Xyl (6) 862.3 30