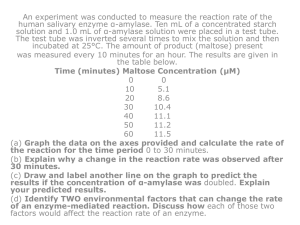
MOLECULAR BIOCHEMISTRY II INTRODUCTORY LECTURE
advertisement
MOLECULAR BIOCHEMISTRY II INTRODUCTORY LECTURE SYLLABUS – AMINO ACID BIOSYNTHESIS – ENERGY METABOLISM OBESITY DIABETES ATKINS DIET – NUCLEOTIDE METABOLISM – DNA STRUCTURE – DNA – PROTEIN INTERACTIONS TRANSCRIPTION FACTORS – DNA METHYLATION – PHOTOSYNTHESIS SOME CHEMICAL PRINCIPLES TO BE COVERED BIOCHEMICAL PATHWAYS – ENZYME CLASSIFICATION – MECHANISMS – REGULATORY CONTROL ROLE OF METAL IONS IN BIOCHEMISTRY PRINCIPLES OF CATALYSIS – TRANSITION STATES COFACTORS – ADDITION OF C1 UNITS OXIDATION/REDUCTION REACTIONS ENZYME CLASSIFICATION SIX CLASSES ( http://us.expasy.org/enzyme/ ) – NOMENCLATURE COMMITTEE OF INTERNATIONAL UNION OF BIOCHEMISTRY AND MOLECULAR BIOLOGY (1992) – COVALENT CHEMICAL BONDS MADE/BROKEN OXIDOREDUCTASES TRANSFERASES HYDROLASES LYASES ISOMERASES LIGASES ADDITIONAL CLASS (“ENERGASES”) – PHYSICAL REACTIONS – NON-COVALENT PRODUCT-LIKE AND SUBSTRATE-LIKE STATES WHAT CONSTITUTES A CHEMICAL BOND? “…there is a chemical bond between two atoms or groups of atoms in case that the forces acting between them are such as to lead to the formation of an aggregate with sufficient stability to make it convenient for the chemist to consider it as an independent molecular species.” Linus Pauling in “The Nature of the Chemical Bond” SIX TRADITIONAL ENZYME CLASSES CAN YOU RECOGNIZE THE CLASS TO WHICH AN ENZYME BELONGS BY LOOKING AT THE OVERALL REACTION? IN-CLASS EXERCISE – FOR THE FOLLOWING 10 REACTIONS WHICH YOU HAVE ALREADY SEEN THUS FAR IN YOUR STUDY OF BIOCHEMISTRY, INDICATE THE ENZYME BY NAME OR BY CLASS SIX ENZYME CLASSES OXIDOREDUCTASE TRANSFERASE HYDROLASE LYASE ISOMERASE LIGASE CATALYSIS OF “PHYSICAL” REACTIONS PRODUCT-LIKE AND SUBSTRATE-LIKE STATES: EXAMPLES : – – – – – – CHAPERONIN-MEDIATED (PROTEIN FOLDING) CHROMATIN CONDENSATION “MOLECULAR MOTOR” OPERATION DNA PROCESSING BY POLYMERASES ACTIVE AND CARRIER-MEDIATED TRANSPORT G-PROTEIN MEDIATED REGULATION OF HORMONE RECEPTORS MEMBRANE TRANSPORTERS (PUMPS) ARE NOW RECOGNIZED AS A SPECIAL CLASS OF ENZYMES “ENERGASES” : TRANSDUCE ENERGY FROM COVALENT BONDS INTO MECHANICAL WORK “ENERGASES” MEDIATE NUCLEOSIDE TRIPHOSPHATE HYDROLYSIS THE FREE ENERGY RELEASED IS COUPLED TO SYSTEM’S CONFORMATIONAL CHANGE ARE ATPases AND GTPases CORRECTLY CLASSIFIED AS “HYDROLASES”? – ATP + H2O ADP + Pi + HEAT Keq = [ADP][Pi] / [ATP] ∆Ghydrolysis IS RELEASED AS HEAT HERE THE ENZYME IS ATPase AND IT’S A HYDROLASE ENERGASE EXAMPLE A SYNTHETASE REACTION: – ATP + GLU + NH3 GLN + ADP + Pi – HERE THE ∆Ghydrolysis IS COUPLED TO ∆Gsynthesis THROUGH A REACTIVE INTERMEDIATE – Keq = [GLN][ADP][Pi] / [ATP][GLU][NH3] = [GLN] / [GLU][NH3] X [ADP][Pi] / [ATP] AN ENERGASE REACTION: – – – – – ATP + STATE 1 + H2O ADP + STATE 2 + Pi HERE THE ∆Ghydrolysis IS COUPLED TO ∆Gconformational change Keq = [STATE 1] / [STATE 2] X [ADP][Pi] / [ATP] NOTICE SIMILARITY TO Keq FOR SYNTHETASE REACTION THERE’S NO CHEMICAL (COVALENT) CHANGE, THOUGH ENZYMES AS MECHANOCHEMICAL PROTEINS THE GIBBS FREE ENERGY OF ATP HYDROLYSIS IS TRANSDUCED INTO A FORM OF USEFUL WORK – TRANSLATION – ROTATION – SOLUTE GRADIENT A RECIPROCAL RELATIONSHIP – ENZYMES USE NON-COVALENT INTERACTIONS TO BREAK COVALENT BONDS – ENERGY FROM BREAKING COVALENT BONDS CAN MODIFY NON-COVALENT INTERACTIONS KEY CONCEPTS IN ORGANIC CHEMISTRY THE “SIX PILLARS” – – – – – – ELECTRONEGATIVITY POLAR COVALENT BONDING STERIC EFFECTS INDUCTIVE EFFECTS RESONANCE AROMATICITY Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 ELECTRONEGATIVITY Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 POLAR COVALENT BONDING Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 STERIC EFFECTS Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 INDUCTIVE EFFECTS Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 RESONANCE Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 AROMATICITY Mullins, J.J. “Six pillars of organic chemistry”, J. Chem. Educ. 2008, 85(1), 83-87 SUGGESTION FOR LEARNING BIOCHEMICAL MECHANISMS WHENEVER POSSIBLE, TRY TO RATIONALIZE MECHANISMS USING ONE OR MORE OF THESE “PILLARS” AN INTRODUCTION TO AMINO ACID METABOLISM NITROGEN CYCLE – THE “FIXTATION” OF NITROGEN THE CENTRAL ROLE OF GLUTAMATE THE NITROGEN CYCLE N2 IS A VERY STABLE MOLECULE – BOND ENERGY = 941.4 kJ/MOL – COMPARED TO 498.7 kJ/MOL FOR O2 – A SINGLE C=O BOND IN CO2 IS 799 kJ/MOL HOW IS IT METABOLIZED (“FIXED”)? THE “NITROGEN CYCLE” – PRODUCTION OF METABOLICALLY USEFUL NITROGEN NITRITES NITRATES AMMONIA THE NITROGEN CYCLE N-FIXING ORGANISMS: – ANAEROBES MARINE CYANOBACTERIA “DIAZOTROPHS” DIAZOTROPHS – COLONIZE ROOT NODULES OF LEGUMES – GENUS Rhizobium SYMBIOTIC RELATIONSHIP – ENZYME IS “NITROGENASE” THE NITROGENASE REACTION: N2 + 8 H+ + 8 e- + 16 ATP + 16 H2O 2 NH3 + H2 + 16 ADP + 16 Pi – REQUIRES ATP AND ELECTRONS – CONTAINS Fe AND Mo THE NITROGEN CYCLE ENERGETICALLY COSTLY – NEED 16 ATPs TO “FIX” ONE N2 MOLECULE COMPARE THIS TO INDUSTRIAL FIXATION: – TEMPERATURE 300o - 500o C – PRESSURE > 300 ATM – METAL CATALYST NH3 FORMED IS USED IN FORMATION OF – GLUTAMATE (Glu Dehydrogenase) – GLUTAMINE (Gln Synthetase) EXCESS NH3 EXCRETED INTO SOIL RESTORE USABLE NITROGEN BY PLANTING ALFALFA THE NITROGEN CYCLE MOST PLANTS DO NOT SUPPORT N-FIXING BACTERIA NEED PRE-FIXED NITROGEN SOURCE – NH3 – NO2– NO3- SOURCES: – LIGHTNING (10% OF NATURALLY-FIXED N) – FERTILIZERS – DECAY OF ORGANIC MATTER IN SOIL THE NITROGEN CYCLE PLANTS, FUNGI, BACTERIA REDUCE NO3-: – A TWO-STEP PROCESS NO3- + 2H+ + 2e- NO2- + H2O – ENZYME: NITRATE REDUCTASE NO2- + 8H+ + 6e- NH4+ + 2H2O – ENZYME: NITRITE REDUCTASE SOME BACTERIA CAN OXIDIZE NH4+ – “NITRIFICATION” – NH4+ NO2- AND THEN TO NO3- DENITRIFICATION – CONVERSION OF NO3- TO N2 BY OTHER BACTERIA THE NITROGEN CYCLE ATMOSPHERIC N2 IS THE ULTIMATE NITROGEN SOURCE DENITRIFICATION N2 NO3NITROGEN FIXATION NITRATE REDUCTASE NITROGENASE NITRITE REDUCTASE NH4+ NO2NITRIFICATION ORGANISMS ASSIMILATE NH3 ROLE OF GLUTAMINE SYNTHETASE – MICRO-ORGANISMS: ENTRY POINT FOR FIXED N – GLU + ATP + NH4+ GLN + ADP + Pi IN ALL ORGANISMS, GLN IS AN AMINO GROUP CARRIER GLUTAMATE SYNTHASE IN BACTERIA, PLANTS – -KETOGLUTARATE + GLN + NADPH + H+ 2 GLU + NADP+ OVERALL RXN’: -KG + NH4+ + ATP + NADPH + H+ GLU + NADP+ + ADP + Pi THE CENTRAL ROLE OF GLUTAMATE “GLUTAMATE FAMILY” OF AMINO ACIDS – DEGRADATIVE METABOLISM CONVERGES ON THAT OF GLU GLU GLN PRO HIS ARG ORNITHINE GLU IS THE PRECURSOR OF – – – PRO ORNITHINE ARG GLU/-KG ARE TRANSAMINATION PARTNERS – AMINO ACID + -KG GLU + -KETOACID OXIDATIVE DEAMINATION OF GLU (GLU DEHYDROGENASE) GLU + NAD(P)+ + H2O -KG + NAD(P)H + NH4+ N-ACETYLGLUTAMATE SYNTHESIS – – ALLOSTERICALLY REGULATES CPS I OF UREA CYCLE GLU + ACETYL-CoA N-ACETYL GLUTAMATE Kelly A., Stanley CA. (2001). “Disorders of Glutamate Metabolism”. Mental RetardAtion and Developmental Disorders. 7:287-295. CLOSING POINTS HIGH ENERGY COSTS TO FIX NITROGEN – ITS USE MUST BE CAREFULLY CONTROLLED GLU AND GLN ARE PIVOTAL IN AMINO GROUP TRANSFER – GLU OFTEN DONATES THE AMINO GROUP – GLN STORES, CARRIES AMINO GROUPS TRANSAMINASES – CATALYSTS FOR TRANSFER OF AMINO GROUPS TO αKETOACIDS – FREELY REVERSIBLE REACTIONS IMPORTANT IN BOTH SYNTHETIC AND DEGRADATIVE PATHWAYS