Lecture 8
advertisement

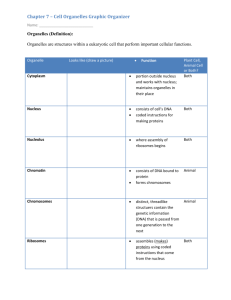
1. Compartmentation of Eukaryotic Cells a. Different intracellular compartments distinct functions i. Cytosol 1. Main compartment of Cytoplasm a. Excludes membrane bound organelles 2. Gelatenous mass a. ~20% protein by mass 3. Cytoribosomes 4. Enzymes 5. Cytoskeleton ii. Membrane Bound organelles 1. Nucleus a. Site of DNA & RNA synthesis b. Envelope has a double membrane structure c. Houses genome 2. Endoplasmic Reticulum (ER) a. Continuous membrane b. Synthesis of lipids and proteins c. Ca regulation d. Rough ER i. Ribosomes on surface (makes it rough) ii. Protein Synthesis e. Smooth ER i. No ribosomes ii. Lipid Synthesis iii. Ca store 3. Golgi Apparatus a. Near nucleus b. Modification, Synthesis and sorting of protein/lipids made in ER c. Vesicles on either side 4. Lysosome a. Contains enzymes for intracellular digestion of macromolecules and organelles b. Garbage collector of the cell 5. Endosome a. Contains and sorts endocytosed material b. Can fuse with Lysosome i. Degradation 6. Vacuole a. Major feature of i. Plant ii. Fungal cells b. Storage or Degradative Functions 7. Mitochondrion a. Double membrane structure b. ATP synthesis c. 1700/cell 8. Chloroplast (plants) a. Carbon fixation by photosynthesis b. Double membrane structure c. Thylakoid Membrane d. ATP synthesis e. CO2 fixation 9. Peroxisome a. Oxidation of toxic molecules b. Fatty acid breakdown b. Relative volume of major intracellular compartments in individual cells i. Membrane-bound organelles 1. ~50% of cytoplasm volume 2. Area and mass of intracellular membrane usually much greater than plasma membrane (12-25x) ii. Relative amounts of organelle types differ in different cells 1. Difference reflect functions of cells in different tissues c. Sorting of proteins to different membrane compartments i. Synthesis of almost all cell proteins begins in the cytosol 1. Mitochondrial proteins dont ii. Fate of protein depends on the sorting signals found in the amino acid sequence 1. If no sorting signal: protein stays in cytosol 2. If sorting signal present: protein goes to specific organelle iii. Specific sorting signals could be required for retention in, or export from, a compartment iv. Signals located in transported vesicle or protein are recognized by receptor protein in target organelles v. Two types of sorting signals 1. Signal peptide (or signal sequence) a. Continuous stretch of amino acids i. Usually at end of polypeptide ii. N’ end b. May be removed by a signal peptidase i. It is a protease c. Experimental manipulation i. Protein with signal sequence in ER ii. Protein with out signal sequence in Cytosol iii. Put signal sequence on protein in cytosol 1. Protein in cytosol moved into ER iv. Protein with signal sequence removed moved into cytosol 1. Signal sequence is sufficient for sorting d. Examples i. Cytosol to 1. ER 2. Chloroplasts 3. Mitochondria 4. Peroxisomes 5. Nucleus ii. Golgi to ER (retention) 2. Signal patch a. Composed of non-continuous segments of amino acid sequence i. Nuclear Localization Sequences b. Identifying NLS i. Comparing a mutated NLS to a wild type nonmutated NLS ii. Identify difference in amino acid sequence 1. Determine which amino acids are necessary for NLS function c. Formation of patch requires protein folding d. Examples i. Nuclear import d. Three main ways proteins move into compartments i. Gated Transport 1. Selective gate: active transport of specific macromolecules; small molecules diffuse freely 2. Example a. Import through nuclear pore ii. Transmembrane Transport 1. Use membrane-bound protein ‘translocators’ 2. Transported protein unfold during transport 3. Example a. Cytosol mitochondria 4. Protein assisted pathway iii. Vesicular transport 1. Membrane “sidedness’ and protein orientations are preserved 2. Example: a. ER Golgi e. Methods i. Autoradiography 1. Allows visualization of biochemical processes 2. Process a. Radioactively labeled b. Specimen fixed for microscopy/autoradiography i. Dipped in liquid emulsion 1. Similar to old cameras 3. Can locate the proteins that are made up by the amino acids that were labeled 4. Pulse Chase experiment a. Pulse i. Brief incubation with radioactivity during which amino acids are incorporated into proteins b. Chase i. Period when the tissue is exposed to the unlabeled medium ii. Additional proteins synthesized using nonradioactive amino acids 5. Secretory pathway a. Technique first defined this pathway ii. Green Fluorescent Protein (GFP) 1. Isolated Jellyfish gene a. Produces GFP proteins i. Emit green light 2. Allows to follow the dynamic movements of specific proteins in the living organism a. Detected by UV microscopy iii. Subcellular Fractionation 1. Vesicles derived from different organelles have different properties a. Allows for separation from one another 2. Fractions identified by electron microscopy iv. Cell-free systems 1. Isolation of a microsomal fraction by differential centrifugation a. Able to see that isolated parts of the cell had remarkable activities b. Homogenate and shove thing down tube 2. RER vesicles trap newly synthesized secreted proteins 3. (Siekevitz,Palade) v. Genetic mutants 1. Genes encode for abnormal proteins a. Unable to carry out typical function b. Characteristic frequency 2. Provides information on the function of the normal protein 3. Yeast secretory pathway established this way