hep26453-sup-0007
advertisement

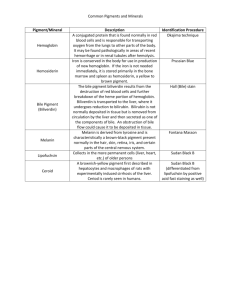
Supporting experimental procedures Bile acid measurement by mass spectrometry Three days after surgery, the liver was collected and aliquots were snap frozen in liquid nitrogen. Bile acids were extracted from frozen liver samples as previously described (1). Briefly, liver samples were cut into small pieces and boiled for 5 minutes in ethanol. Liver extracts were filtered, mixed with 2 µl of internal standard solution and incubated with 0.5 mol/L ammonium carbonate for 30 minutes at 60°C. Samples were then submitted to solidphase extraction on reverse-phase Chromabond C18 cartridges (100 mg; Macherey-Nagel, Düren, Germany). Bile acids were dissolved in methanol and injected into HPLC-MS/MS. Chromatographic separation was performed with an Agilent Technologies 1100 HPLC apparatus, in series with the turbo ion spray source of the mass spectrometer QTRAP 2000 (Applied Biosystems-Sciex, Framingham, MA). The data were acquired with Analyst software, version 1.4.2 (Applied Biosystems-Sciex). Immunoprecipitation and silver staining Total liver proteins (1 mg) were pre-cleared with A/G Plus-agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C and incubated with anti-E-cadherin (5 µg) overnight at 4°C. New A/G Plus-agarose beads were added and the incubation continued for 2 h. Immunoprecipated proteins were resuspended in Laemmli buffer, boiled and subjected to SDS-PAGE. The gel was then fixed in 50% ethanol, 5% acetic acid and washed in 50% ethanol for 1 h each. After sensitization in 0.02% sodium thiosulfate, the gel was silver stained in cold 0.1% silver nitrate for 20 min at 4°C and developped in 0.05% formalin, 3% sodium carbonate. Protein bands of interest were excised from gel and destained for 15 minutes in 15 mmol/L potassium ferricyanide and 50 mmol/L sodium thiosulfate. Following 2 washes with water, the gel pieces were dehydrated in 100% acetonitrile for 15 minutes and finally washed with water. Excised bands were stored dry at -20°C until mass spectrometry analysis. Identification of proteins by mass spectrometry In-gel digestion was carried out with trypsin as described by Bussone et al (2) and using a Freedom EVO 100 digester/spotter robot (Tecan, Männedorf, Switzerland) for all steps. After treatments, the sample analysis was achieved with a chromatographic separation using an Ultimate 3000 (Dionex, Sunnyvale, CA) series high performance liquid chromatography (HPLC). Fractions were then spotted online on a MALDI target using a Probot (Dionex) fraction collector. Sample analysis by mass spectrometry was performed with a MALDITOF-TOF 4800 mass spectrometer (Applied Biosystems, Carlsbad, CA) as previously described (2). Database searching was carried out using Mascot version 2.2 (MatrixScience, London, UK) combining MS and MS/MS interrogations on mouse proteins from Swissprot databank (www.expasy.org). Supporting references 1. Debray D, Rainteau D, Barbu V, Rouahi M, El Mourabit H, Lerondel S, et al. Defects in gallbladder emptying and bile Acid homeostasis in mice with cystic fibrosis transmembrane conductance regulator deficiencies. Gastroenterology 2012;142:1581-1591 e1586. 2. Bussone G, Dib H, Dimitrov JD, Camoin L, Broussard C, Tamas N, et al. Identification of target antigens of self-reactive IgG in intravenous immunoglobulin preparations. Proteomics 2009;9:2253-2262.