5.1.1 Cellular Control

OCR A2 UNIT F215 CELLULAR CONTROL
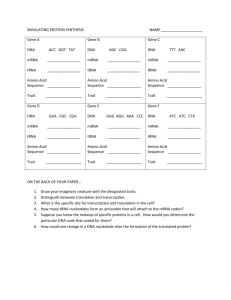
Specification a) State that genes code for polypeptides, including enzymes b) Explain the meaning of the term genetic code c) Describe. with the aid of diagrams, the way in which a nucleotide sequence codes for the amino acid sequence in a polypeptide d) Describe. with the aid of diagrams, how the sequence of nucleotides within a gene is used to construct a polypeptide, including the roles of messenger
RNA, transfer RNA and ribosomes e) State that mutations cause changes to the sequence of nucleotides in DNA molecules f) Explain how mutations can have beneficial, neutral or harmful effects on the way a protein functions g) State that cyclic AMP activates proteins by altering their three-dimensional structure h) Explain genetic control of protein production in a prokaryote using the lacoperon i) Explain that the genes that control development of body plans are similar in plants, animals and fungi, with reference to homeobox sequences j) Outline how apoptosis (programmed cell death) can act as a mechanism to change body plans
1
Definitions
Word to define Definition
Gene A length of DNA (sequence of nucleotides) that codes for one polypeptide. A gene is part of a DNA molecule (one chromosome is one DNA molecule)
Polypeptide
*Note that the text book definition is incorrect
A polymer consisting of a chain of amino acids joined by peptide bonds
Genome
Protein
The entire DNA sequence of an organism. The human genome consists of about 25000 genes and 3 billion nucleotide base pairs.
In eukaryotic cells, most genes are on the nuclear chromosomes.
A few are in the mitochondria or chloroplasts
A large polypeptide (usually 100 or more amino acids). Some proteins contain one polypeptide chain; some contain more than one polypeptide chain
Types of Polypeptides/Proteins coded for by Genes
Structural proteins such as collagen
Haemoglobin
Antibodies (immunoglobulins)
Cell surface receptor proteins
Hormones such as insulin and glucagon
Regulatory proteins involved in switching genes on/off
Actin and myosin in muscle cells
Tubulin in microtubules
Channel proteins
Electron carriers
Enzymes*
Enzymes are very important proteins because they control metabolic processes in the cell, including the synthesis of non-protein molecules in cells
2
The Genetic Code
The genetic code is the genetic information that codes for the assembly of amino acids into polypeptides and proteins
Features of the genetic code
The genetic code is a triplet code . This means that each amino acid in the polypeptide is coded for by a sequence of three DNA bases. A triplet of bases is called a codon
There are 4 different bases in DNA. When these are arranged in groups of three, the number of different triplet sequences is 4 3 = 64. Since there are 20 different amino acids in proteins, 64 triplet codes is more than enough to code for 20 amino acids
The genetic code is degenerate . This means that all amino acids except methionine have more than one codon coding for them. Most of this degeneracy involves the third nucleotide in the codon. One implication is that a mutation that substitutes one DNA base for another may not alter the amino acid coded for
Some codons are stop codons. They do not code for an amino acid and their presence in the DNA sequence indicates the end of the polypeptide chain
The genetic code is non-overlapping. Each DNA base is part of the codon for only one amino acid
Non-overlapping Code: C G T A G A
(2 separate triplets, 2 amino acids coded for)
Overlapping Code : C G T A G A
CGT
GTA
TAG
AGA
(4 overlapping triplets, 4 amino acids coded for)
If the code was overlapping, more information could be stores in the same space but the code would be less flexible since each codon would depend partly on the bases of the previous codon
3
The genetic code is widespread but not universal. All organisms on Earth share the same genetic code except for a few exceptions
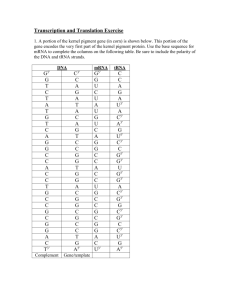
Tables of the genetic code can be given in terms of DNA and mRNA
The Genetic Code in terms of DNA
Use the DNA-amino acid table above to list all the DNA codons that code for each amino acid listed in the table below. The left hand column gives the first base. The top line gives the second base and the right hand column gives the third base
Amino Acid DNA codons that code for this amino acid
Ala Alanine
Arg Arginine
Asn Asparagine
Asp Aspartic acid
Cys Cysteine
Gln Glutamine
Glu Glutamic acid
Gly Glycine
His Histidine
Iso Isoleucine
GCC, GCT,
GCA, GCG
No. of codo ns
4
Amino Acid
Leu leucine
Lys lysine
Met methionine
Phe phenylalanine
Pro proline
Ser serine
Thr threonine
Try tryptophan
Tyr tyrosine
Val valine
DNA codons that code for this amino acid
No. of codons
4
The Genetic Code in terms of mRNA
Use the mRNA-amino acid table above to list all the mRNA codons that code for each amino acid listed in the table below.
Amino Acid
Ala Alanine
Arg Arginine
Asn Asparagine
Asp Aspartic acid
Cys Cysteine
Gln Glutamine
Glu Glutamic acid
Gly Glycine
His Histidine
Iso Isoleucine mRNA codons that code for this amino acid
GCC, GCU,
GCA, GCG
No. of codo ns
4
Amino Acid
Leu leucine
Lys lysine
Met methionine
Phe phenylalanine
Pro proline
Ser serine
Thr threonine
Try tryptophan
Tyr tyrosine
Val valine mRNA codons that code for this amino acid
No. of codons
5
The figure below is a circular representation of the genetic code
Answer the following questions:
1. Does the circular diagram above represent the genetic code in terms of DNA or mRNA?
…………………………………………………………………………………………
2. Where in the circle would you look for the following? (use descriptions such as middle, 1 st concentric ring and 2 nd concentric ring)
the first base in the codon
…………………………………………………………………………………………
the second base in the codon
…………………………………………………………………………………………
the third base in the codon
…………………………………………………………………………………………
6
3. Write out the following codons (remember there could be more than one codon)
Stop codons
………………………………………………………………………………………….
Glycine
…………………………………………………………………………………………..
Threonine
………………………………………………………………………………………….
4. List the amino acid sequence of the following sequence of codons:
AUGCGGUAUUUUGGAAAAGUUUGA
………………………………………………………………………………………….
5. Write an alternative codon sequence for the same amino acid sequence:
………………………………………………………………………………………….
6. Write out the DNA sequence that was transcribed to produce the mRNA strand given in question 4
………………………………………………………………………………………….
7. What is the feature of the genetic code that allows different base sequences in mRNA and DNA to code for the same amino acid sequence?
…………………………………………………………………………………………
7
The table below shows a comparison of the nucleotides in cDNA and the amino acid sequences for a protein called crystalline in the lens of the mammalian eye for three mammals
Explain why there is a higher similarity between the amino acid sequences than between the cDNA sequences (cDNA stands for copy DNA)
……………………………………………………………………………………………….
……………………………………………………………………………………………….
……………………………………………………………………………………………….
……………………………………………………………………………………………….
………………………………………………………………………………………………
………………………………………………………………………………………………
Stages in Polypeptide Synthesis
Transcription
This is the first stage of polypeptide synthesis that occurs in the nucleus
Definition of Transcription
The formation of a single stranded mRNA copy of the DNA coding strand
Requirements for mRNA synthesis
The gene to be transcribed
Free RNA nucleotides (stored in the nucleolus). The activated RNA nucleotides are ATP, GTP, CTP and UTP
RNA polymerase
8
Events in Transcription
1. A gene unwinds and unzips (catalysed by RNA polymerase). This occurs as the gene region dips into the nucleolus. Unzipping involves the breaking of hydrogen bonds between complementary base pairs of the two DNA strands
2. Only one DNA strand is copied into single stranded mRNA
3. Activated RNA nucleotides line up against the exposed complementary bases on the template strand of the DNA section. Hydrogen bonds form between the complementary bases: U binds to A, C to G and A to T
4. RNA polymerases catalyses the formation of phosphodiester bonds between the RNA nucleotides. The two extra phosphate groups of each RNA nucleotide are hydrolysed to release energy for phosphodiester bond formation
5. The mRNA produced is complementary to the template DNA strand of the gene but is a copy of the coding DNA strand of the gene
6. The mRNA is released from the DNA and passes out of the nucleus, through a nuclear pore in the nuclear envelope, to a ribosome in the cytoplasm
Complete the table below:
DNA coding strand
DNA template strand mRNA
Amino acid*
UUA AUG CGU
*use the genetic code to complete the amino acid row
Translation
GGA UAA0
Definition of Translation
Translation is the second stage of polypeptide synthesis when the amino acids are assembled into a polypeptide at a ribosome in the cytoplasm
The mRNA code consists of codons of 3 bases. The base/codon sequence in the mRNA determines the amino acid sequence
9
Revision of Ribosomes and tRNA
Ribosomes
Assembled in the nucleolus of eukaryote cells from ribosomal RNA (rRNA) and protein
Each ribosome has two subunits – one large and one small. The mRNA fits into a groove in the small subunit tRNA
tRNA molecules are made in the nucleus. They pass into the cytoplasm via the nuclear pores
tRNA is a single stranded length of RNA that folds into a clover leaf shaped structure
At one end, three unpaired bases bind a specific amino acid. At the other end, three unpaired bases form the anticodon that binds temporarily to the mRNA codon during translation
The function of tRNA molecules is to bring specific amino acids to the ribosome so that peptide bonds can be formed between the amino acids as a polypeptide is synthesised
Events during Translation
1. A molecule of mRNA fits in the groove of the ribosome. Two codons (6 bases) attach to the small ribosomal subunit
2. The first mRNA codon is always AUG – the start codon that codes for the amino acid methionine. A tRNA molecule carrying methionine has the anticodon UAC. This anticodon is complementary to the mRNA codon AUG.
Anticodon and codon bind as hydrogen bonds form between the complementary bases
3. A second tRNA carrying a different amino acid binds to the second exposed codon by complementary base pairing between codon and anticodon
4. A peptide bond forms between the two adjacent amino acids, catalysed by an enzyme in the small subunit
5. The ribosome now moves along the mRNA to the third codon. When this occurs, the mRNA slides through the ribosomal groove.
10
6. A third tRNA brings another amino acid and a peptide bond forms between the third amino acid and the dipeptide. The first tRNA leaves the ribosome and is available to collect another amino acid molecule in the cytoplasm
7. The polypeptide chain continues to be assembled until a stop codon on the mRNA is reached.
Role of Cyclic AMP in Activating Proteins cAMP activates proteins by changing their 3D shape to make them complementary to the molecule they bind to.
Reference was made to cAMP in F214 when its role in activating enzymes was described during the hormonal action of adrenaline on target cells
In muscle cells, the enzyme glycogen phosphorylase that hydrolyses glycogen to glucose, is activated by cAMP. cAMP binds to an allosteric site on glycogen phosphorylase so that its active site shape is more complementary and exposed to the glycogen substrate for efficient binding.
Protein Synthesis in Prokaryotes
Since the DNA is not contained within a nucleus, translation begins as soon as mRNA has been synthesised
Mutations
Definitions
A mutation is a random change in the nucleotide sequence of a DNA molecule
Mutagens , such as ionising radiation (X-rays, UV radiation) and certain chemicals
(eg benzopyrene in cigarette smoke) increase the chance of mutation
There are two categories of mutations
1. A chromosome mutation involves changes to parts or of whole chromosomes
2. A DNA/gene mutation is a change in the nucleotide base sequence within a gene
11
There are several types of DNA/gene mutation :
Substitution point mutations in which one base pair replaces another. If a substitution occurs in the first base pair of a triplet , it is likely to change one amino acid in the primary structure of a polypeptide. Changes to the second or third base are less likely to change the amino acid – these are silent mutations
Insertion and deletion mutations in which one or more nucleotide pairs are inserted or deleted from a length of DNA. These cause a frameshift
Stutter mutations where triplets are repeated . Huntington’s disorder is caused by a stutter mutation
Frameshift – this is a change in the reading frame of the coding DNA template and mRNA so that the amino acid sequence changes after the mutation. Frameshifts can have major effects on the polypeptide produced
– it may not function or be longer or shorter if a stop codon is removed or added
The effects of a deletion causing a frameshift mutation on a sequence of five amino acids
Coding DNA strand …. A T G T A C G G C T T A C G T T A G …….
Template DNA strand
…. T A C A T G C C G A A T G C A A T C ……. mRNA strand
…. A U G U A C G G C U U A C G U U A G…… amino acids met tyr gly leu arg stop
Effects of deletion of the fifth base pair
Coding DNA strand
…. A T G T C G G C T T A C G T T A G …….
Template DNA strand
….T A C A G C C G A A T G C A A T C ……. mRNA strand … A U G U C G G C U U A C G U U A G ….. amino acids met ser ala tyr val ser/arg
Note that deletion of one base pair alters the amino acid sequence after the mutation
12
Effects of Mutations and Examples
Consequences of
Mutation
Harmful mutations
Neutral mutations
Description
The amino acid sequence of the polypeptide is changed such that the protein no longer functions correctly
These have no effect on the fitness or survival of the organism
Useful mutations These produce alleles that are beneficial to the survival of the organism.
The alleles may be useful if there are changes in the environment
Examples/Types of mutation
Huntington disease due to a stutter mutation in the gene coding for
Huntington’s protein. The gene contains too many CAG sequences.
The symptoms of this disease include dementia and loss of motor control later in life
Cystic fibrosis 70% of cases are due to the deletion of a triplet of base pairs from the gene coding for normal polypeptide
Sickle cell anaemia results from a point mutation on codon 6 of the gene coding for the β-polypeptide of haemoglobin. Valine replaces glutamic acid in the polypeptide chain
Tumours due to a point mutation in proto-oncogenes - these genes are growth promoting genes. A point mutation changes the genes into oncogenes – they remain switched on permanently and cause unregulated cell division
Mutant triplet codes for the same amino acid (silent mutation)
Mutation occurs in non-coding regions of DNA and is not expressed in the phenotype
Triplet codes for a different amino acid but this makes no difference to the polypeptide function or gives no advantage or disadvantage to the organism eg some people have free ear lobes, some have attached
Antibiotic resistance in members of a bacterial population
The possession of one sickle cell anaemia allele in heterozygotes confers protection against the malarial parasite
13
The genetic control of protein production in a prokaryote using the lac operon
An operon is a group of genes that act together to control a biochemical pathway. The theory of the lac operon was described by Jacob and Monod in
1961. They received the Nobel prize for their research into the genetic control of protein production involved in lactose metabolism in Escherichia coli bacteria.
Background
Enzymes involved in basic cellular functions such as glycolysis are synthesised at a fairly constant rate
Inducible enzymes are synthesised at varying rates, according to environmental changes within the cell
Bacteria normally respire glucose but if there is no glucose available and lactose is present in the cell, they can respire lactose
The lac operon
The lac operon is a group of genes on a section of DNA that act together to produce the three enzymes required for lactose metabolism:
1. β galactosidase that catalyses lactose hydrolysis to glucose and galactose
2. Lactose permease that transports lactose into the cell
3. Another enzyme
Lactose is the inducer of the lac operon
Diagram showing the components of the lac operon and its regulator gene which is some distance away
14
Components of the lac operon and its regulator gene
Component of Functions
lac operon
Structural genes Z codes for β galactosidase that hydrolyses lactose
Y codes for lactose permease that increases uptake of lactose
A codes for another enzyme
All structural genes code for mRNA during transcription and code for the synthesis of enzymes
Operator gene
Promoter gene
Length of DNA next to the structural genes
Where repressor protein binds when there is no lactose in the medium
Does not code for a polypeptide
It can switch the structural genes on or off
Region of DNA to which RNA polymerase binds to begin transcription of Z,
Y and A
Does not code for a polypeptide
Regulator gene Not part of the operon region of DNA and some distance away
Codes for a repressor protein that has two binding sites, one for lactose when lactose is present in the medium and one for the operator region when there is no lactose in the medium
The regulatory gene controls the expression of the structural genes
When there is no lactose in the medium, the repressor protein product of the regulator gene switches the structural genes off. It does this by binding to the operator region preventing RNA polymerase binding with the promoter region.
15
How the lac operon works
Lactose absent from the growth medium
1 Regulator gene is expressed and the repressor protein is synthesised
Lactose present in the growth medium
1 Regulator gene is expressed and the repressor protein is synthesised
Lactose binds to the repressor protein causing the repressor protein to change shape
2 The repressor protein binds to the operator region, covering the promoter region where RNA polymerase normally binds
2 Repressor protein can no longer bind to the operator region and dissociates from it
5 Without mRNA, β galactosidase, lactose permease and the third enzyme cannot be synthesised
4 Benefits – energy and amino acids are saved
3 The promoter region is unblocked.
RNA polymerase can now bind to it allowing transcription of mRNA
4 for genes Z, Y and A
3 mRNA molecules for genes Z, Y and A are translate d to produce β galactosidase, lactose permease and the third enzyme
5 E.coli
bacteria can use lactose permease to take up lactose into the cells and β galactosidase to hydrolyse lactose to glucose and galactose. These two monosaccharides are respiratory substrates
Negative and positive control of the lac operon
Negative control by the repressor protein
Glucose is the preferred respiratory substrate. When glucose is present, the repressor protein binds to the operator gene and this prevents transcription of the structural genes. The repressor protein has a negative effect on structural gene transcription
Positive control by cyclic AMP receptor protein (CRP)
Even when lactose is available and no glucose, RNA polymerase does not bind readily to the promoter region of the operon. The protein CRP is needed to help the
16
binding of RNA polymerase to promoter region. CRP requires the attachment of cyclic AMP to change its 3D shape and expose its DNA binding site. Once bound to
DNA, the activated CRP helps RNA polymerase to bind to promoter to start transcription – activated CRP has a positive effect on structural gene transcription
Question
A strain of the bacterium Escherichia coli has been discovered that has a mutation in the regulator gene. One aspect of the phenotype of this mutant is that it produces large concentrations of β galactosidase at all times
1) Define the term phenotype
A phenotype is an observable characteristic of an individual due to interaction of its genotype with the environment.
2) Explain why a mutation in the regulator gene leads to a constant production of β galactosidase
The mutation was alter the DNA of the gene therefore there will be a change in how the regulatory gene would be expressed. Therefore, the repressor protein would no longer be complementary to the promoter region so RNA polymerase will bind to the promoter region. Then the operator gene would
‘turn on’ the structural genes and the enzymes beta-galactosidase and lactose permease would be created. There would be constant production due the promoter region being free from the repressor protein. It is possible that the repressor protein may not even be produced due to the change in DNA and change in AA sequence.
Answer:
Mutation causes alteration of the DNA of the regulator gene
Therefore repressor protein mRNA transcribed is altered
Either no repressor protein is synthesised or the protein has a different amino acid sequence
Structure and shape of the repressor protein could change
17
Such that the repressor protein is unable to bind to the operator region
The promoter region can bind to RNA polymerase
Transcription and translation of the structural genes continues
Homeobox genes controlling body plans
Definition of homeobox genes
Homeobox genes control the early development of the body plan of animals, plants and fungi. This control gives the pattern to the body including the polarity (positions of the head and tail), the segmentation pattern of insects and mammals and positioning of the organs.
Studies of the control of animal development
The fruit fly, Drosophila melanogaster , the zebra fish, Danio rerio and the mouse,
Mus musculus , have all been used by scientists to find out more about how genes control animal development. These organisms are ‘model organisms’
The information obtained from studying ‘model organisms’ can be applied to humans because:
1 Humans are in the same animal kingdom
2 All animals have eukaryotic cells
3 They have genes in common
4 They have common ancestors
5 They have similar embryonic development patterns
Characteristics of organisms chosen by scientists for studying gene control of development
1 Organisms have short life cycles and therefore mature and develop quickly
2 They are easy to breed, producing many offspring
3 Cheap to buy
4 Are common and readily available
5 Genome has already been sequenced as they have previously been well studied and many mutants are already known
Development of Drosophila melanogaster – the fruit fly
18
The genetic control of cell differentiation has been studied in the fruit fly, Drosophila melanogaster , an invertebrate animal
Drosophila development – see diagrams in textbook page 114
1. The single celled eggs undergo mitotic divisions at a very fast rate (less than
10mins each division)
2. No new plasma membranes are formed initially (no cytokinesis) and a multinucleated syncytium forms
3. After the 8 th division the nuclei migrate to the edges of the cell and by the 11 th division they form an outer layer around the central yolk
4. The division rate slows and the nuclear genes are transcribed
5. The plasma membrane invaginates around the 6000 nuclei forming an outer layer of 6000 cells
6. After 2-3 hours, the embryo divides into a series of segments
– these segments form the body plan of the fruit fly
7. 3 segments merge to produce the head. There are 3 thoracic segments (T1-
3) and 8 abdominal segments (A1-8)
8. When the larval form becomes the adult during metamorphosis, legs, wings and antennae appendages develop
19
20
Genetic control of Drosophila development
The development process described is controlled genetically by homeobox genes
Some genes determine which end is the head (anterior end) and which end is the tail (posterior end). This is referred to the embryo ’s polarity
The segmentation genes determine the polarity of each segment
Homeotic selector genes control the development of each individual segment
- these are the master genes
There are two sets of these master genes:
- The complex that controls development of thorax and abdominal segments
- The complex that controls development of head and thoracic segments
Mutations of these genes can change one body part to another. One example is antennapedia, where the antennae of Drosophila look more like legs
Genetic control of development in other organisms
Homeobox genes work in similar ways in other invertebrate animals, vertebrate animals (such as humans), plants and fungi
Features of the homeobox genes
Homeobox genes are homeotic or regulatory genes that contain homeobox nucleotide sequences
Each gene contains a sequence of 180 base pairs, producing a polypeptide containing 60 amino acids
Some of the polypeptides are transcription factors that bind to genes further along the DNA (upstream genes), starting transcription of the upstream genes. These transcription factors therefore control the expression of other genes, switching them on
The homeobox genes are arranged in clusters called Hox clusters. The more
Hox clusters an organism has, the more complex the organism is
- Roundworms (nematodes) have one Hox cluster
- Drosophila has two Hox clusters
- Vertebrates have four clusters of 9-11 genes on separate chromosomes
21
The increase in number of the Hox clusters probably derives from duplication of the single Hox cluster as found in the Nematodes
The homeobox genes are expressed in specific patterns in certain stages during embryo development, controlling body plan development and the differentiation of embryonic cells
Why homeobox genes are so similar across widely different species
Homeobox genes are vital to the development of an organism
Mutation would alter the body plan and have huge effects on the organism’s development
Mutations are likely to be lethal and selected against
Retinoic acid and birth deformities
Retinoic acid, a derivative of vitamin A is a morphogen that controls the pattern of tissue development
Retinoic acid activates homeobox genes in the same order that they are expressed, directing embryo development from head to tail
The concentration of dietary vitamin A is crucial. If pregnant women
(particularly in the first month of gestation) consume too much vitamin A the expression of these genes is abnormal resulting in birth defects. This is why pregnant women are now advised not to eat liver (liver stores vitamins A and
D)
Apoptosis ( pronounced apo-tosis)
Definition of apoptosis
Apoptosis is programmed cell death , an orderly process that occurs naturally in multicellular organisms. Normal body cells are not immortal and undergo a maximum number of about 50 mitotic divisions (the Hayflick limit). After this, the cells undergo a sequence of controlled biochemical processes to kill the cell
Necrosis is uncontrolled cell death that occurs during tissue infection or damage by trauma such as during a heart attack. The cell death is disorderly and associated with leakage of hydrolytic enzymes from cells that cause tissue damage and inflammation
22
Sequence of events in cells during apoptosis
1. Protease enzymes from lysosomes hydrolyse the cell’s cytoskeleton
2. Cell cytoplasm is dense with tightly packed organelles
3. Changes in the plasma membrane lead to the formation of small bulges called
‘blebs’
4. Chromatin in the nucleus condenses and the nuclear envelope breaks down.
DNA breaks down into fragments
5. The cell divides into separate membrane bound vesicles (the blebs start this process)
6. Phagocytes engulf these vesicles by phagocytosis
7. The useful cell molecules are recycled and the cellular waste products are removed safely so that they do not damage other cells/tissues. No hydrolytic enzymes are released into the tissues
23
Control of apoptosis
Control is complex and involves a network of both intracellular and extracellular cell signalling molecules that trigger an ordered sequence of changes in the cytoplasm
Cytokines secreted by T helper cells, hormones and nitric oxide are some of these molecules
Nitric oxide makes the inner mitochondrial membrane more permeable to hydrogen ions and this makes the proton gradient less steep
– reducing ATP synthesis
Functional living cells produce proteins that inhibit apoptosis. During cell death, other proteins are synthesised within the cell that bind to these inhibitory proteins. Apoptosis can then occur
The extracellular cell signalling molecules bind to receptors on the cell plasma membrane promoting a cascade of events in the cell including the activation of caspases that cause the breakdown of cytoskeleton proteins
Examples of apoptosis
Apoptosis is an important part of plant and animal tissue development. Too many cells are produced during development and some of these are removed by apoptosis
During development of the foot and hand, the finger and toe digits are separated by apoptosis. Apoptosis will be controlled by some Hox genes.
Mutations can cause this apoptosis to be incomplete so that the hands/feet are partially webbed
24
Apoptosis is also important throughout life. 20-30 billion cells per day undergo apoptosis in children between 8 and 14 years. In adults, 50-70 million cells apoptose. The rate of cells dying should equal the rate of cell production
Apoptosis is important when female mammals menstruate – the inner lining of the uterus is broken down
Killer T lymphocytes kill virus infected cells by inducing their apoptosis. After the killer T cells have fulfilled this function they must induce apoptosis in each other to prevent destruction of healthy body cells. Defects in this process result in an auto-immune response and destruction of healthy body tissues, such as in rheumatoid arthritis and multiple sclerosis
Apoptosis occurs in plants to remove cells infected with viruses
When a tadpole develops into a frog, apoptosis causes the tail to be lost
What happens if apoptosis rate is too low?
If mutations occur in the DNA controlling apoptosis, the cells continue to replicate and form a tumour . The rate at which new cells are produced by mitosis and cytokinesis is greater than the rate of cell death.
If some tumour cells break away and are transported in blood or lymph fluids, secondary cancers can occur in other parts of the body. These are called metastases and the cancerous tumour is malignant.
The human papilloma virus (HPV) causes genital warts. This virus can be transmitted sexually and can cause tumour formation in the female cervix. HPV interferes with a protein required for apoptosis.
25