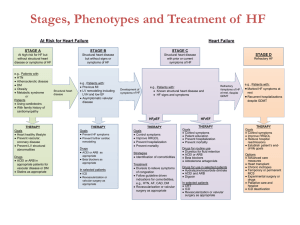
Supplemental Content - JACC: Heart Failure
advertisement

Online Appendix Detailed Statistical Methods A.1. Models We utilized Bayesian hierarchical models to combine data from all trials while accounting for the variation of survival times (or rehospitalization indications) within and between studies. One important feature of the Bayesian hierarchical models is that, because they allow for borrowing of information across studies/subgroups, they allow us to make inferences about subgroups that may be under-represented in a portion of the trials. Moreover, missing values are treated as additional variates whose values can be naturally imputed as part of the estimation procedure (Schafer (1996), and Little and Rubin(1987)). The Bayesian paradigm requires specification of both the likelihood for the observations as well as priors for the model parameters. Preliminary analysis of the survival data from all trials indicated that the Weibull parametric survival model adequately fit the survival data, in particular, with results similar to those obtained under the Cox-Proportional Hazards Model. We thus utilized the Weibull parametric survival model to time to death from all causes. Under this model, we formulate the hazard function for an individual 𝑖 observed in study 𝑠 as: ℎ𝑖 (𝑡) = ℎ0𝑠 (𝑡)𝑒𝑥𝑝(𝜏𝑠 𝑍𝑖 + 𝛾𝑠 𝑊𝑖 + 𝜈𝑍𝑖 𝑊𝑖 + 𝛽′𝑋𝑖 ) (1) where ℎ0𝑠 (∙) is the baseline hazard function in study 𝑠. We assumed that ℎ0𝑠 (𝑡) = 𝑎𝑠 𝑡 𝑎𝑠 −1 , 𝑏𝑠 𝑎𝑠 that is, the hazard function from a Weibull distribution with shape parameter 𝑎𝑠 and scale parameter 𝑏𝑠 . Furthermore, 𝑍𝑖 refers to treatment, 𝑊𝑖 for comorbidity count, and 𝑋𝑖 the vector with the remaining covariates for subject 𝑖. We are interested in investigating whether the efficacy of ICD relative to control is modified by comorbidities and thus we also include an interaction between treatment and comorbidity. The above formulation allows for trial-specific 1 baseline hazard functions, as well as trial-specific main effects of treatment and comorbidities. To allow for borrowing of information, we further assumed that: [𝑎𝑠 |𝜇𝑎 , 𝜎2𝑎 ] ~ LogNormal(𝜇𝑎 , 𝜎2𝑎 ), [𝑏𝑠 |𝜇𝑏 , 𝜎2𝑏 ] ~ LogNormal(𝜇𝑏 , 𝜎2𝑏 ), [𝜏𝑠 |𝜇𝜏 , 𝜎2𝜏 ] ~ Normal(𝜇𝜏 , 𝜎2𝜏 ), [𝛾𝑠 |𝜇𝛾 , 𝜎2𝛾 ] ~ Normal (𝜇𝛾 , 𝜎2𝛾 ). (2) We completed the model formulation with the following priors: [𝜇𝑎 , 𝜇𝑏 , 𝜇𝜏 , 𝜇𝛾 , 𝜈, 𝛽] ∝ 1, (3) that is, we assumed non-informative priors for the population (mean) parameters. Moreover, we assumed that for the variance components, [𝜎𝑎2 ] ~ 𝐼𝑛𝑣𝑒𝑟𝑠𝑒𝐺𝑎𝑚𝑚𝑎(𝑐, 𝑑), [𝜎𝑏2 ] ~ 𝐼𝑛𝑣𝑒𝑟𝑠𝑒𝐺𝑎𝑚𝑚𝑎(𝑐, 𝑑), [𝜎𝜏2 ] ~ 𝐼𝑛𝑣𝑒𝑟𝑠𝑒𝐺𝑎𝑚𝑚𝑎(𝑐, 𝑑), [𝜎𝛾2 ] ~ 𝐼𝑛𝑣𝑒𝑟𝑠𝑒𝐺𝑎𝑚𝑚𝑎(𝑐, 𝑑). In our applications, we utilized 𝑐 = 2, 𝑑 = 1. These values were chosen to be minimally informative and such that, the preliminary analysis of simpler Bayesian hierarchical models (e.g. only random effects for treatment effect or only trial-specific baseline hazard functions), resulted in similar inference to those from the frequentist Cox-proportional hazards model. We formulated a model for the indicator of at least one rehospitalization by analogy. Let 𝜋𝑖 denote the probability that individual 𝑖 had at least one rehospitalization. Then, we assumed 𝑙𝑜𝑔𝑖𝑡(𝜋𝑖 ) = (𝛼𝑠 + 𝜏𝑠 𝑍𝑖 + 𝛾𝑠 𝑊𝑖 + 𝜈𝑍𝑖 𝑊𝑖 + 𝛽′𝑋𝑖 ) (4) with a hierarchical formulation similar to that described by equations (2)-(3). Variations of the above models were formulated to address specific sensitivity analysis. All models were estimated using Markov Chain Monte Carlo (MCMC) methods with WinBUGS. A sample of code written to analyze the data is available in Appendix C. 2 A.2. Missing Data Let 𝐾𝑗𝑖 denote the indicator of comorbid condition 𝑗 in individual 𝑖. To allow for imputation of missing values, we assume that each comorbid condition follows a Bernoulli model. Specifically, let 𝑞𝑗 denote the probability of presence of comorbid condition 𝑗. This probability is assumed known and based on clinical assumptions from the trial populations. That is, for trials missing certain comorbidities, overall rates for the trial could be extrapolated from similar trials. The MADIT I and MADIT II patients in our cohort are very similar (ischemic/CAD, EF 35-40% cutoff) and could be used to impute rates of comorbid disease in each-other's populations. The ischemic subgroup in SCD-HeFT (52%) could be added to this group. In contrast, patients in the DEFINITE trial were similar to the non-ischemic group in SCD-HeFT (48%), and thus overall rates of disease in these populations could inform one another. Therefore, we assume that 𝐾𝑗𝑖 ~𝐵𝑒𝑟𝑛𝑜𝑢𝑙𝑙𝑖 (𝑞𝑗 ). Each comorbid condition is imputed via simulation prediction within each iteration of the MCMC procedure. Thus the procedure incorporates the uncertainty on the missing value of the comorbid condition. For sensitivity analysis, we also consider the extreme situations where the uncertainty is removed by assuming 𝑞𝑗 = 0 (best case – subject with missing data does not have condition) or 𝑞𝑗 = 1 (worst case – subject with missing data has condition). Finally, we note that in model (1), the comorbidity count is 𝑊𝑖 = ∑𝑗 𝐾𝑗𝑖 . Thus, for individuals with any missing comorbidity, the comorbidity count varies according to the imputed comorbidity values. A3. Posterior Inference Under the Weibull proportional hazards and logistic regression models (1)-(4) there are natural measures of treatment effectiveness as given by the hazard ratio and odds ratio, respectively. In addition, we consider absolute measures of treatment effectiveness. Specifically, we calculate the risk differences (RD) and, its reciprocal, the number needed to treat (NNT). We 3 use the procedure proposed by Austin (2010) for logistic regression which naturally extends to the proportional hazards model. We note that inferences are all within the Bayesian approach. The advantage is that it allows us to obtain posterior credible intervals for these measures without relying on asymptotic results or on bootstrap methods. In this Section we describe the procedure. Specifically, under the survival model, we calculate risk differences using survival probabilities at given times. Specifically, to compute the risk differences in the survival model, we determine the survival probability at a given year 𝑡 post-randomization in the hypothetical situation when all subjects have 𝑤 comorbidities and are treated with ICD. Denote the subjectspecific survival probabilities as 𝑆𝑖1 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 1). Likewise, we determine survival probability at a given year 𝑡 post-randomization in the hypothetical situation when all subjects have 𝑤 comorbidities but are not treated with ICD. The corresponding survival probabilities are 𝑆𝑖0 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 0). These probabilities are derived from the model specified in equation (1). Note that these probabilities depend on covariates 𝑋𝑖 . The risk difference, based on survival probabilities at year 𝑡 post-randomization when all subjects have 𝑤 comorbidities is computed by integrating out those probabilities with respect to the distribution of the covariates. Using the empirical distribution of the covariates in the study and Monte Carlo integration this leads to the risk difference: 𝑅𝐷(𝑡|𝑊 = 𝑤) = 1 0 ∑𝑁 𝑖=1 𝑆𝑖 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 1) − 𝑆𝑖 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 0). 𝑁 The inverse of the above quantity gives the number needed to treat. Note that this quantity also depends on the number of comorbidities. We can also find a marginal risk difference by averaging over the empirical distribution of the comorbidities and obtain: 𝑅𝐷(𝑡) = 1 0 ∑𝑁 𝑖=1 𝑆𝑖 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤𝑖 , 𝑍𝑖 = 1) − 𝑆𝑖 (𝑡|𝑋𝑖 , 𝑊𝑖 = 𝑤𝑖 , 𝑍𝑖 = 0). 𝑁 4 By analogy, under the logistic regression model we compute the probability of rehospitalization when all subjects have 𝑤 comorbidities and are treated with ICD, that is, 𝜋𝑖1 (𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 1). Likewise, we determine probability of rehospitalization when all subjects have 𝑤 comorbidities but are not treated with ICD. The corresponding survival probabilities are 𝜋𝑖0 (𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 0). These probabilities are derived from equation (4). Thus, in the logistic regression model we obtain the risk difference when all subjects have 𝑤 comorbidities as: 𝑅𝐷(𝑤) = 1 0 ∑𝑁 𝑖=1 𝜋𝑖 (𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 1) − 𝜋𝑖 (𝑋𝑖 , 𝑊𝑖 = 𝑤, 𝑍𝑖 = 0). 𝑁 We can also find a marginal risk difference by averaging over the empirical distribution of the comorbidities and obtain: 1 0 ∑𝑁 𝑖=1 𝜋𝑖 (𝑋𝑖 , 𝑊𝑖 = 𝑤𝑖 , 𝑍𝑖 = 1) − 𝜋𝑖 (𝑋𝑖 , 𝑊𝑖 = 𝑤𝑖 , 𝑍𝑖 = 0). 𝑅𝐷 = 𝑁 References Little RJA and Rubin DB. (2002). Statistical Analysis with Missing Data. Wiley-Interscience; 2 edition. Schafer (1997). Analysis of Incomplete Multivariate Data. Chapman & Hall. Chapman and Hall/CRC; 1 edition. Austin PC (2010). Absolute risk reductions, relative risks, relative risk reductions, and numbers needed to treat can be obtained from a logistic regression model. Journal of Clinical Epidemiology, 63: 2-6. 5 Table 1. Definitions of comorbidities by trial. Smoking Ischemic heart disease Chronic kidney disease MADIT-I MADIT-II DEFINITE SCD-HeFT Any cigarette smoking prior to randomization Any cigarette smoking prior to randomization History of current or past smoking History of current or past smoking Prior MI, as inclusion for trial Prior MI, as inclusion for trial Only non-ischemic cardiomyopathy included (significant ischemic disease was an exclusion) Patients with ischemic cardiomyopathy, prior MI, prior CABG, or prior PCI Calculated eGFR (<60 mL/min/1.73 m2) by the used the Chronic Kidney Disease Epidemiology Collaboration equation.* Diabetes History of insulin-dependent diabetes History of diabetes History of insulin dependent or noninsulin dependent diabetes History of diabetes Pulmonary disease Interstitial pulmonary findings on x-ray Not available History of COPD History of pulmonary disease Not available Not available History of atrial fibrillation and/or flutter, either paroxysmal or chronic History of atrial fibrillation or atrial flutter History of peripheral vascular disease Not available History of peripheral vascular disease Not available Atrial fibrillation Peripheral vascular disease Unless noted, definitions are as gathered and stated on the original trial case report form. Further diagnostic details are not available. *Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. May 5 2009;150(9):604-612. MADIT-I: Multicenter Automatic Defibrillator Implantation Trial I; MADIT-II: Multicenter Automatic Defibrillator Implantation Trial II; DEFINITE: Defibrillators in Non-Ischemic Cardiomyopathy Treatment Evaluation; SCD-HeFT: Sudden Cardiac Death in Heart Failure Trial; MI: myocardial infarction; CAD: coronary artery disease; CABG: coronary artery bypass grafting; PCI: percutaneous coronary intervention; eGFR: estimated glomerular filtration rate; COPD: chronic obstructive pulmonary disease; 6 Table 2. Distribution of number of comorbidities, by treatment assignment. No. of comorbidities Control (n=1577) ICD (n=1771) 0 69 84 1 319 358 2 569 673 3 450 459 4 149 173 5 20 23 6 1 1 7 0 0 7 Table 3. Baseline characteristics stratified by number of observed comorbidities Trials MADIT-I MADIT-II DEFINITE SCD-HeFT Treatment Assignment Control ICD Age Female Race White Black Other Medical History Hypertension Ischemic Heart Disease Prior CABG Medical Therapy Beta-Blocker ACE Inhibitor Antiarrhythmic Therapy LVEF LBBB <2 Comorbidities ≥2 Comorbidities P-value (n=830) (n=2518) <0.001 22 (3) 157 (6) 74 (9) 1015 (40) 248 (30) 210 (8) 486 (59) 1136 (45) 0.841 388 (47) 1189 (47) 442 (53) 1329 (53) <0.001 55 (13) 62.4 (10.6) <0.001 254 (31) 425 (17) <0.001 587 (71) 2078 (83) 189 (23) 318 (13) 54 (7) 122 (5) 268 (46) 132 (16) 68 (12) 1271 (55) 1918 (76) 1088 (47) <0.001 <0.001 <0.001 624 (75) 746 (90) 14 (2) 22.7 (6.7) 172 (30) 1605 (64) 2143 (85) 92 (4) 23.6 (6.3) 390 (18) <0.001 <0.001 0.004 0.001 <0.001 Baseline characteristics, co-morbidities, treatment assignment, and medical therapy, stratified by number of comorbidity. Values are presented as number (%) or mean (standard deviation). ICD: implantable cardioverter-defibrillator; MADIT-I: Multicenter Automatic Defibrillator Implantation Trial I; MADIT-II: Multicenter Automatic Defibrillator Implantation Trial II; DEFINITE: Defibrillators in Non-Ischemic Cardiomyopathy Treatment Evaluation; SCD-HeFT: Sudden Cardiac Death in Heart Failure Trial; CABG: coronary artery bypass grafting; ACE: angiotensinconverting enzyme; LVEF: left-ventricular ejection fraction; LBBB: left bundle branch block. 8 Table 4. Unadjusted outcomes by comorbidity. <2 Comorbidities (n=830) ≥2 Comorbidities (n=2518) Control (n=388) ICD (n=442) Control (n=1189) ICD (n=1329) 63 (16) 42 (10) 351 (30) 270 (20) Cardiac, arrhythmic 25 (40) 5 (12) 128 (37) 59 (22) Cardiac, non-arrhythmic 8 (13) 13 (31) 64 (18) 70 (26) Cardiac, NOS 7 (11) 2 (5) 46 (13) 49 (18) Non-cardiac 9 (14) 9 (21) 61 (17) 62 (23) Unknown 14 (22) 13 (31) 52 (15) 30 (11) Rehospitalization 142 (54) 192 (63) 629 (62) 853 (73) ICD complication 0 (0) 57 (18) 5 (1) 249 (21) 0 (0) 10 (3) 0 (0) 23 (2) Death Infection Values are presented as number (%); cause-specific mortality presented as percentages of all deaths. ICD complications are described in the Appendix, and only apply to ICD patients and those in the control group who crossed-over. ICD: implantable cardioverter-defibrillator; NOS: not otherwise specified. 9 Table 5. All missing comorbidities imputed to true (patient has disease). No. of comorbidities Log HR for ICD vs. Control Absolute Survival Difference at 5 years (ICD – Control) Median 95% CI Median 95% CI 0 -1.26 (-2.15, -0.35) 0.10 (0.05, 0.16) 1 -1.08 (-1.91, -0.27) 0.12 (0.06, 0.17) 2 -0.91 (-1.69, -0.12) 0.13 (0.07, 0.18) 3 -0.73 (-1.48, 0.01) 0.13 (0.08, 0.17) 4 -0.56 (-1.27, 0.18) 0.11 (0.07, 0.15) 5 -0.38 (-1.08, 0.38) 0.08 (0.02, 0.14) 6 -0.21 (-0.89, 0.58) 0.04 (-0.05, 0.13) posterior probability of interaction between treatment and comorbidity, p=0.002. HR: hazard ratio; ICD: implantable cardioverter-defibrillator; CI: confidence interval. 10 Table 6. All missing comorbidities imputed to false (patient does not have disease). No. of comorbidities Log HR for ICD vs. Control Absolute Survival Difference at 5 years (ICD – Control) Median 95% CI Median 95% CI 0 -0.94 (-1.79, -0.15) 0.13 (0.06, 0.19) 1 -0.77 (-1.54, -0.04) 0.13 (0.08, 0.19) 2 -0.60 (-1.31, 0.05) 0.13 (0.08, 0.17) 3 -0.43 (-1.14, 0.26) 0.10 (0.05, 0.15) 4 -0.26 (-0.99, 0.46) 0.06 (-0.03, 0.14) 5 -0.08 (-0.86, 0.7) 0.00 (-0.13, 0.12) 6 0.09 (-0.76, 0.96) -0.05 (-0.2, 0.09) posterior probability of interaction between treatment and comorbidity, p=0.02. HR: hazard ratio; ICD: implantable cardioverter-defibrillator; CI: confidence interval. 11