bit25697-sup-0001-SuppData-S1
advertisement

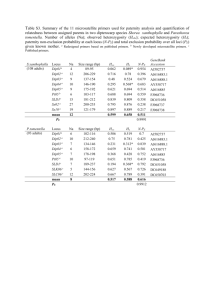
Supporting Information for: Directing positional specificity in enzymatic synthesis of bioactive 1- phosphatidylinositol by protein engineering of a phospholipase D Jasmina Damnjanović, Chisato Kuroiwa, Hidetoshi Tanaka, Ken Ishida, Hideo Nakano and Yugo Iwasaki* Laboratory of Molecular Biotechnology, Graduate School of Bioagricultural Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan Running title: Engineering of 1-PI specific phospholipase D Keywords: phospholipase D, 1-phosphatidylinositol, positional specificity Figure S1. Outline of the overlapping PCR used to introduce single residue mutations. A 1.8 kbp fragment was amplified using primers CATR1 and one of the four reverse primers (NYR-186X-Rv, NYR-188X-Rv, NYR-189X-Rv or NYR-190X-Rv listed in Table SI) with nyr as a template. Another 1.0 kbp fragment was amplified using one of the specific forward primers listed in Table SI and T7T with nyr as a template. The two fragments were joined by overlapping PCR using primers, CATR1 and T7T to afford the full length PLD gene of 2.8 kbp containing single residue mutation. Figure S2. Analysis of the protein-ligand interactions in crystal structure of Streptomyces antibioticus PLD (PDB: 2ZE9). Ligand: 1,2-diheptanoyl-sn-glycero-3-phosphate. 3-PI (%) 1-PI (%) Figure S3. 1-PI/3-PI ratio produced by NYR-186X/189X double mutants, as determined by LC-MS analysis. As a reference, parent NYR produces 76% of 1-PI and 24% of 3-PI. Figure S4. SDS-PAGE of the purified enzyme fractions (10% gel): lane M- molecular weight marker (Takara, Japan) followed by lanes of the purified NYR and its variants. Figure S5. Presentation of the polar interactions, including hydrogen bonds, (yellow dots) formed between myo-inositol and residues of the active site and acceptor-binding site after docking of inositol to the phosphatidyl-bound model structures of NYR and GYR. Ptd denotes phosphatidyl moiety while myo-Ins denotes myo-inositol. Backbone of inositol and phosphatidyl moiety is colored light blue while, that of the residues is colored green. Oxygen atoms are colored red, hydrogen atoms white and nitrogen atoms dark blue. Phosphorus is presented in orange color. Figure S6. Analysis of hydrogen bond network formed by residue 188 in model structures of NYR (K188), NYR-188W, NYR-188E and NYR-188N. Hydrogen bonds are colored yellow. Red, blue and magenta boxes indicate main chain-main chain, side chain-side chain and main chain-side chain interactions, respectively. Table SI. List of the primers used in this study. Primer Name Sequence (5’-3’) a) Generation of G186X variants NYR-G186A ACGGGCGGGATCAACGCCAACAAGGACGACTA NYR-G186C ACGGGCGGGATCAACTGCAACAAGGACGACTA NYR-G186D ACGGGCGGGATCAACGACAACAAGGACGACTA NYR-G186E ACGGGCGGGATCAACGAGAACAAGGACGACTA NYR-G186F ACGGGCGGGATCAACTTCAACAAGGACGACTA NYR-G186K ACGGGCGGGATCAACAAGAACAAGGACGACTA NYR-G186H ACGGGCGGGATCAACCACAACAAGGACGACTA NYR-G186I ACGGGCGGGATCAACATCAACAAGGACGACTA NYR-G186L ACGGGCGGGATCAACCTGAACAAGGACGACTA NYR-G186M ACGGGCGGGATCAACATGAACAAGGACGACTA NYR-G186N ACGGGCGGGATCAACAACAACAAGGACGACTA NYR-G186P ACGGGCGGGATCAACCCGAACAAGGACGACTA NYR-G186Q ACGGGCGGGATCAACCAGAACAAGGACGACTA NYR-G186R ACGGGCGGGATCAACCGCAACAAGGACGACTA NYR-G186S ACGGGCGGGATCAACTCGAACAAGGACGACTA NYR-G186T ACGGGCGGGATCAACACCAACAAGGACGACTA NYR-G186V ACGGGCGGGATCAACGTCAACAAGGACGACTA NYR-G186W ACGGGCGGGATCAACTGGAACAAGGACGACTA NYR-G186Y ACGGGCGGGATCAACTACAACAAGGACGACTA NYR-G186X-Rv GTTGATCCCGCCCGCCCGT b) Generation of K188X variants NYR-K188A GGGATCAACGGCAACGCGGACGACTACCTCGAC NYR-K188C GGGATCAACGGCAACTGCGACGACTACCTCGAC NYR-K188D GGGATCAACGGCAACGACGACGACTACCTCGAC NYR-K188E GGGATCAACGGCAACGAGGACGACTACCTCGAC NYR-K188F GGGATCAACGGCAACTTCGACGACTACCTCGAC NYR-K188G GGGATCAACGGCAACGGGGACGACTACCTCGAC NYR-K188H GGGATCAACGGCAACCACGACGACTACCTCGAC NYR-K188I GGGATCAACGGCAACATCGACGACTACCTCGAC NYR-K188L GGGATCAACGGCAACCTGGACGACTACCTCGAC NYR-K188M GGGATCAACGGCAACATGGACGACTACCTCGAC NYR-K188N GGGATCAACGGCAACAACGACGACTACCTCGAC NYR-K188P GGGATCAACGGCAACCCGGACGACTACCTCGAC NYR-K188Q GGGATCAACGGCAACCAGGACGACTACCTCGAC NYR-K188R GGGATCAACGGCAACCGCGACGACTACCTCGAC NYR-K188S GGGATCAACGGCAACTCGGACGACTACCTCGAC NYR-K188T GGGATCAACGGCAACACGGACGACTACCTCGAC NYR-K188V GGGATCAACGGCAACGTGGACGACTACCTCGAC NYR-K188W GGGATCAACGGCAACTGGGACGACTACCTCGAC NYR-K188Y GGGATCAACGGCAACTACGACGACTACCTCGAC NYR-K188X-Rv GTTGCCGTTGATCCCGCCCG c) Generation of D189X variants NYR-D189A ATCAACGGCAACAAGGCCGACTACCTCGACACC NYR-D189C ATCAACGGCAACAAGTGCGACTACCTCGACACC NYR-D189E ATCAACGGCAACAAGGAGGACTACCTCGACACC NYR-D189F ATCAACGGCAACAAGTTCGACTACCTCGACACC NYR-D189G ATCAACGGCAACAAGGGCGACTACCTCGACACC NYR-D189H ATCAACGGCAACAAGCACGACTACCTCGACACC NYR-D189I ATCAACGGCAACAAGATCGACTACCTCGACACC NYR-D189K ATCAACGGCAACAAGAAGGACTACCTCGACACC NYR-D189L ATCAACGGCAACAAGCTGGACTACCTCGACACC NYR-D189M ATCAACGGCAACAAGATGGACTACCTCGACACC NYR-D189N ATCAACGGCAACAAGAACGACTACCTCGACACC NYR-D189P ATCAACGGCAACAAGCCGGACTACCTCGACACC NYR-D189Q ATCAACGGCAACAAGCAGGACTACCTCGACACC NYR-D189R ATCAACGGCAACAAGCGCGACTACCTCGACACC NYR-D189S ATCAACGGCAACAAGTCGGACTACCTCGACACC NYR-D189T ATCAACGGCAACAAGACCGACTACCTCGACACC NYR-D189V ATCAACGGCAACAAGGTCGACTACCTCGACACC NYR-D189W ATCAACGGCAACAAGTGGGACTACCTCGACACC NYR-D189Y ATCAACGGCAACAAGTACGACTACCTCGACACC NYR-D189X-Rv CTTGTTGCCGTTGATCCCGC d) Generation of D190X variants NYR-D190A ACGGCAACAAGGACGCGTACCTCGACACCG NYR-D190C ACGGCAACAAGGACTGCTACCTCGACACCG NYR-D190E ACGGCAACAAGGACGAGTACCTCGACACCG NYR-D190F ACGGCAACAAGGACTTCTACCTCGACACCG NYR-D190G ACGGCAACAAGGACGGCTACCTCGACACCG NYR-D190H ACGGCAACAAGGACCACTACCTCGACACCG NYR-D190I ACGGCAACAAGGACATCTACCTCGACACCG NYR-D190K ACGGCAACAAGGACAAGTACCTCGACACCG NYR-D190L ACGGCAACAAGGACCTCTACCTCGACACCG NYR-D190M ACGGCAACAAGGACATGTACCTCGACACCG NYR-D190N ACGGCAACAAGGACAACTACCTCGACACCG NYR-D190P ACGGCAACAAGGACCCGTACCTCGACACCG NYR-D190Q ACGGCAACAAGGACCAGTACCTCGACACCG NYR-D190R ACGGCAACAAGGACCGCTACCTCGACACCG NYR-D190S ACGGCAACAAGGACTCGTACCTCGACACCG NYR-D190T ACGGCAACAAGGACACGTACCTCGACACCG NYR-D190V ACGGCAACAAGGACGTCTACCTCGACACCG NYR-D190W ACGGCAACAAGGACTGGTACCTCGACACCG NYR-D190Y ACGGCAACAAGGACTACTACCTCGACACCG NYR-D190X-Rv GTCCTTGTTGCCGTTGATCCC e) Sequencing and common primers pld-seq1 TGGCTGCTGCACACCCCCGGCT pld-seq2 ATCTACCACCTCAACGTGGTGC pld-seq3 ACCCGGTGTCGGACGTGGACATGG pld-seq4 ACCCCTCCTCGGGATACCACCCGGA pld-seq5 CGCCGGGTCAAGGTCCGCATCG T7P TAATACGACTCACTATAGGG T7T GCTAGTTATTGGTCAGCGG CATR1 TAGCAGATCTGAGCTCACTAGTGGATCCTCGAATTTCTGCCATTCAT