supporting informations6
advertisement

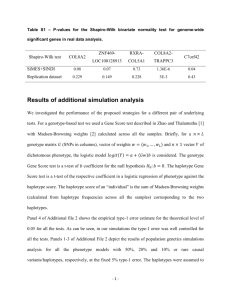
SUPPORTING INFORMATIONS6 Title: Genetic landscape with sharp discontinuities shaped by complex demographic history in moose (Alces alces) Authors: Lovisa Wennerström, Nils Ryman, Jean-Luc Tison, Anna Hasslow, Love Dalén, and Linda Laikre Affiliation of first and senior authors (LW, NR, LL): Division of Population Genetics, Department of Zoology, Stockholm University, SE-106 91 Stockholm, Sweden E-mail adresses: lovisa.wennerstrom@zoologi.su.se,nils.ryman@popgen.su.se, linda.laikre@popgen.su.se, Journal: Journal of Mammalogy mtDNA haplotypes We found a total of 7 haplotypes when sequencing the first hypervariable part of the mitochondrial DNA control region (CR1) comprising a total of 568 (haplotypes 1-6) or 643 (haplotype 7) nucleotides in 44 moose collected in Sweden representing the major genetic groupings (Northern cluster n=17 individuals, Southern cluster n=20, and the transition zone between them divided into a northern (n=4) and a southern part (n=3). The variation in length is due to a 75 base pair (bp) insertion (indel) found in 1 of the individuals. This insertion represents an extra indel occurring in addition to a 75 bp indel found in European moose (including all of the haplotypes of the current dataset) and in some Asian haplotypes, but not in North American moose (Mikko and Andersson 1995; Hundertmark et al 2002; Niedziałkowska et al. 2014). Both 75 bp insertions constitute a repetitive sequence known as RS2 (Douzery and Randi 1997); after excluding it a total of 15 polymorphic sites, all transitions, were found among the haplotypes identified. Haplotype 1 was found in Norrbotten and corresponds to a previously described haplotype in Finland (AF412231; Hundertmark et al. 2002). Haplotype 2 corresponds to the previously described Swedish haplotype (AF412230; Hundertmark et al. 2002) and constitutes the most common haplotype in our samples, found in 35 individuals, corresponding to a frequency of 0.80. Haplotypes 1, 2, and 3 have also been observed by Niedziałkowska et al. (2014). Haplotype 7 has not been reported previously. When excluding the extra 75 bp indel of haplotype 7 the remaining sequence of this haplotype corresponds to the sequence of haplotype 2. Thus, haplotype 7 is the same sequence as haplotype 2 but with the 75 bp indel being “doubled”. The sequence of the “doubled” 75 bp indel of haplotype 7 is the same as the “normal” 75 bp indel of haplotypes 2, 4 and 5. 1 Fig. S6.1. Median joining network of haplotypes 1-6 mtDNA haplotypes found in 44 analyzed Swedish moose. We used POPART (http://popart.otago.ac.nz) to construct the network. The haplotypes are colored according to the different populations identified by STRUCTURE using our 15 nuclear markers. The size of the haplotype is reflecting the number of individuals. Hatch-marks represent mutations, the black circle represents an inferred missing haplotype. Haplotype 7 with the extra 75 bp indel is not included here. 2 Fig. S6.2. Phylogenetic tree for the sequence variation of the first hypervariable part of the mitochondrial DNA control region (CR1) in Swedish moose constructed using MRBAYES v3.2.2 (Ronquist et al. 2012). The tree was constructed using the best fitting model of nucleotide evolution (HKY) inferred using jModeltest 2.1.2 (Darriba et al. 2012).To ensure convergence before discarding 10% as burn-in of 1.1 Million generations, two independent runs were performed. Haplotypes representing the same CR1 mtDNA region as we investigated here were obtained from GenBank. We used a total dataset containing i) our 44 control region sequences, ii) 4 haplotypes from Finland (GenBank accession numbers AF412231-AF412234; Hundertmark et al 2002), iii) 1 haplotype from Sweden (GenBank accession number AF412230; Hundertmark et al. 2002), iv) 17 haplotypes from Europe (GenBank accession numbers KJ831595-KJ831611; Niedziałkowska et al. 2014),and v) as outgroups moose (Alces alces) from Kazakstan (GenBank no JN632595) and reindeer 3 (Rangifer tarandus; Genbank no AB245426). It was not possible to resolve haplotypes 2, 5, 6, and 7 to different branches. The posterior probabilities of nodes are shown. Literature cited (not provided in the paper) DARRIBA, D., G. L. TABOADA, R. DOALLO, D. POSADA. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9:772-772. RONQUIST, F., M. TESLENKO, P. VAN DER MARK, D. AYRES, A. DARLING, S. HÖHNA, B. LARGET, L. LIU, M. A. SUCHARD, AND J. P. HUELSENBECK. 2011. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 3:539-542. 4