Haplotype Inferences with the ELB Algorithm
advertisement

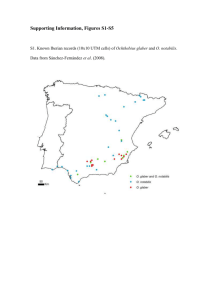
Haplotype Inferences with PHASE v.2 The three-marker haplotypes frequency distributions for LG2 and LG12 showed a large number of unique haplotypes and very similar frequencies of shared haplotypes (Table S4-1). Two-marker haplotypes, used to estimate haplotypic LD, were obtained by collapsing the three-marker haplotypes from either side (LG2 and LG12) or by direct inference (LG10). Shared haplotype frequencies increased in a similar way for Qp46-Qr87, Qr8-Qr112 and Qr112-Qr30, while Qp119-Qp46 departed from the previous pattern with almost twice the number of unique haplotypes. The haplotypes frequency distribution in LG10 (Qr11-Qr96) showed the lowest number of unique haplotypes, but resembled rather closely those from both LG12 and from the second LG2 segments. All segments showed rather high haplotype probabilities (Figure S4-1), which are a function of the data homozygosity and of the coalescent based approach implemented in PHASE (Stephens et al., 2001). Shared haplotypes had slightly higher probabilities than unique haplotypes (Figure S4-1 left panel), as expected because of the use of the coalescent approach. On the other hand, the small probability differences shown by the two species (Figure S4-1 right panel) could be explained by the single marker heterozygosities. Anyway, median haplotype probabilities were very high in all three LGs (LG2: 0.78; LG10: 0.96; and LG12: 0.76). Note that collapsing the three-marker into two-marker haplotypes logically increases the corresponding probabilities. 1 Haplotype Inferences with the ELB Algorithm (using ARLEQUIN v.3.5.1) The ELB algorithm (Excoffier et al., 2003) is a Bayesian method for reconstructing the (unknown) gametic phase of multilocus genotypic data. A detailed description of the method is available within ARLEQUIN's manual (http://cmpg.unibe.ch/software/arlequin3). The comparison with the haplotype reconstructions made with PHASE (Table S4-2) showed that the ELB algorithm identified a slightly larger number of unique haplotypes than PHASE, which might be a consequence of the coalescent-based approach. However, both methods identified the same frequent haplotypes. The inferred haplotypes probabilities were lower with the ELB algorithm than with PHASE (Figures S4-1,2), again reflecting differences in the coalescent-based approach. Finally, the two methods similarly identified few haplotypes shared between the two oak species (Figure S4-3 and Figure 3). In both instances, LG 10 was an exception to this rule, as its most frequent haplotype was common to both species. In spite of the differences between the two haplotype reconstruction methods, the final outcomes regarding haplotypic LD were very much alike (Table 3 and Table S5-3). Methods S4 Methods for haplotype reconstructions using PHASE have been described in the main text and in Supplemental File 3. Haplotype reconstructions from multilocus genotype data, using the ELB algorithm, were carried out with Arlequin v.3.5.1 (Excoffier et al., 2010). 2 The Dirichlet prior (alfa value), the weights given to haplotypes differing by a single mutation from present haplotypes (epsilon value) and the parameter preventing adaptive windows to grow too much (gamma value) were those recommended for SSRs (0.01, 0.1 and 0 respectively). The heterozygote site influence zone was allowed to include all markers from each linkage group (3 markers for LG 2, LG9 and LG 12 and 2 markers for LG 10). The burn-in steps in the Gibbs sampler were set up to the maximum allowed value (9999999). We then obtained 20.000 gametic phases which were sampled every 1.000 iterations. The gametic phases with highest posterior probabilities from each LG were selected to represent the true haplotypes. Figures S4-2 and S4-3 from this supplemental file were prepared with the library “Rcomander” (Fox et al., 2010) under the R environment (R Development Core Team, 2010). References S4 Excoffier L, Laval G, Balding D (2003) Gametic phase estimation over large genomic regions using and adaptive window approach. Human Genomics, 1 (1), 7-19 Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetic analyses under Linux and Windows. Molecular Ecology Resources (in press). Fox J, with contributions from Liviu Andronic, Michael Ash, Theophilius Boye, Stefano Calza, Andy Chang, Philippe Grosjean, Richard Heiberger, G. Jay, Kerns, Renaud Lancelot, Matthieu Lesnoff, Uwe Ligges, Samir Messad, Martin Maechler, Robert Muenchen, Duncan Murdoch, Erich Neuwirth, Dan Putler, Brian Ripley, Miroslav Ristic and Peter Wolf. (2010). Rcmdr: R Commander. R package version 1.5-5. http://CRAN.R-project.org/package=Rcmdr R Development Core Team (2010). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org. 3 Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics, 68, 978-989. 4 Table S4-1 Frequency distributions of the haplotypes inferred by PHASE. Two-marker haplotypes (Bi-H1 and Bi-H2) were obtained after collapsing the respective three-marker haplotypes (Tri-H) from both sides. K is the number of different haplotypes and N is the total number of haplotypes. Haplotype Linkage Group 2 Frequencies Tri-H Bi-H1 Bi-H2 1 2 3 4 5 6 7 8 9 10 11 13 14 16 17 18 19 20 21 24 27 30 55 204 37 15 3 0 1 1 0 1 0 K N 262 356 129 35 24 6 3 1 1 1 1 0 60 33 14 10 1 2 1 2 0 2 Linkage Group 9 LG10 Linkage Group 12 Tri-H Bi-H1 Bi-H2 Bi-H Tri-H Bi-H1 Bi-H2 114 20 1 3 0 0 0 0 0 0 51 25 8 3 3 0 1 0 1 0 87 24 5 3 0 1 0 0 0 0 41 21 13 8 4 3 2 4 0 1 1 2 200 38 12 4 0 0 1 0 0 0 1 59 28 12 6 3 3 2 2 2 0 1 70 24 6 3 6 3 3 1 0 1 1 2 1 1 1 1 1 2 1 2 1 1 1 1 1 202 356 136 356 138 168 5 92 168 120 168 104 368 256 346 122 346 119 346 Table S4-2: Comparison between the haplotype frequencies estimated by PHASE and the ELB algorithm. LG2, LG9 and LG12 show the three-marker haplotypes frequencies, while LG 10 shows the two-marker haplotypes frequencies. LG 2 Haplotype Frequencies Phase 1 2 3 4 5 6 7 8 9 10 11 13 14 16 17 18 19 20 21 24 27 30 55 204 37 15 3 0 1 1 0 1 0 K N 262 356 LG 9 LG 10 ELB Phase ELB Phase 218 37 13 2 0 1 0 0 1 0 114 20 1 3 0 0 0 0 0 0 124 15 2 2 0 0 0 0 0 0 41 21 13 8 4 3 2 4 0 1 1 2 LG 12 ELB 46 21 10 10 3 2 1 5 0 0 1 1 2 Phase ELB 200 38 12 4 0 0 1 0 0 0 1 212 34 6 3 5 0 0 0 0 0 1 256 346 261 346 1 1 1 1 2 1 1 272 356 138 168 143 168 6 104 368 106 368 Figure S4-1: Probabilities assigned by PHASE to the inferred haplotypes. The distributions compare the probabilities for All vs. Shared haplotypes (left panel) and for the haplotypes that belong to each of the two species (right panel). LG2 LG9 LG10 LG12 All Shared Q. robur Q. petraea 7 Figure S4-2: Haplotype probabilities frequency distributions for the best reconstructions with the ELB algorithm. 8 Figure S4-3: Within-species frequency distributions for the best haplotype reconstructions with the ELB algorithm. 9