Deep sequencing of paired colorectal cancers and metastases
advertisement

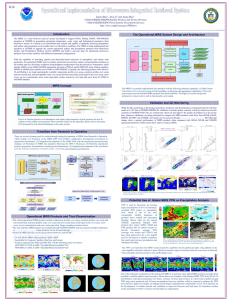
Deep sequencing of paired colorectal cancers and metastases: Detection of novel miRs and colonization specific miRs M. Neerincx1, T.E. Buffart1, D.L.S. Sie2, M.A. van de Wiel3, H. Dekker1, B. Diosdado2, P.P. Eijk2, N.C.T. van Grieken2, G.A. Meijer2, H.M.W. Verheul1 1 Dept. Medical Oncology VU University Medical Center 2 Dept. Pathology VU University Medical Center 3 Dept. Epidemiology and Biostatistics VU University Medical Center INTRODUCTION: Biomarkers are increasingly used for selecting the most optimal therapy for each patient. MiRs have recently been recognized as promising candidate biomarkers because of their role in cancer biology while being protected from degradation. It is unknown whether miRs are differentially expressed between primary colorectal cancers (CRC) and metastatic lesions. Such a comparison may provide information on the potential use of miRs for therapy selection. Moreover, the full miR-ome of CRC has not been elucidated and consequently miR expression have not been studied in a thoroughly manner. AIMS: 1 )Elucidate the miRNA expression of known and potential novel candidate miRs in mCRC and 2) Compare miR expression of primary CRCs and metastases from the same patients. METHODS: RNA was isolated from 64 snap frozen resection specimens, including 16 primary CRCs with 17 corresponding metastases. All tumor samples contained >70% tumor cells. Next generation sequencing was performed on the Illumina Highseq 2000 platform. Sequence data were aligned to miRBase v19. Prediction of novel candidate miRs was based on specific folding characteristics of the precursor sequences and fit of sequenced RNAs to the model of miRNA biogenesis. Cluster analysis and differential expression analysis for known and novel miRs were performed using EdgeR in a paired manner. RESULTS: 622.183.482 small RNA sequences were obtained for 64 samples, identifying 1634 known and 401 potential novel unique mature or complementary strand miRs expressed in mCRC. Unsupervised clustering showed a trend of close relationship between samples of the same patient, indicating that expression profiles of metastases resemble those of primary CRCs. 10 miRs were upregulated and 9 miRs were downregulated (FDR <0.1) in the metastases compared to their paired primary tumors. Including one novel candidate miR and miRs known in metastasis formation (miR-10b), tumor suppression (miR-133, miR-486) or associated with worse prognosis (miR-199b). Interestingly, subgroup analysis revealed specific expression changes for different organs of metastases, indicating that these miRs might contribute to organ specific colonization. CONCLUSION: Using NGS, 401 potential novel miRs were identified in mCRC. MiR expression profiles of primary CRCs resemble those of their metastases. Only 1.15% of the miRs may be differentially expressed between primary CRCs and corresponding metastases, which is most likely determined by organ specificity. Based on our results, we expect that miR expression can be used as a biomarker for therapy selection irrespective of a primary or secondary origin of the CRC tissue.