Investigating microRNA control of nodulation in single cell types
advertisement

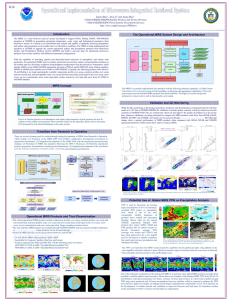
Investigating microRNA control of nodulation in single cell types Explain the overall aim of the project, including the biological question being addressed To improve food security we need to gain an in-depth understanding of how the environment controls plant development. To deal with low nitrogen levels in the soil, leguminous plants form a symbiosis with nitrogen fixing Rhizobia soil bacteria in root nodules. If we can understand nodulation we could transfer it to our major non-legume crops such as rice or wheat with massive benefit. To do this we must identify the molecular mechanisms controlling how a nodule is formed but also, critically for transferability, pinpoint molecules that recognise the bacteria as a compatible symbiont activating genes to switch on development of a nodule, rather than a pathogen, activating expression of defense genes. To identify master regulators of symbiosis we are currently generating single root cell type timeseries of microarray expression data in Medicago (a legume) as it responds during nodulation and during lateral root development responses to nitrogen influx. As well as transcription factors we know from our own previous work in Arabidopsis (Gifford et al 2008 PNAS) and from work in other groups (e.g. Zhou et al 2011 Genes & Development) that microRNAs (miRs) play many major regulatory roles in root responses to the environment, acting in a highly cell-specific manner. In particular miRs have recently been found to regulate specific defense genes and thus might be involved in differentiating defense and symbiosis responses (Shivaprasad et al 2012 Plant Cell). This project aims to identify miRs as master regulators of nodulation and predict whether they could be used to develop nodulation outside legumes. What biological techniques will the student use? Fluorescence-Activated Cell Sorting (FACS) is used to isolate single cell types followed by whole genome expression profiling using microarrays and the student will be trained in these 'omic techniques since we have all facilities for these within our lab. As well as generating new FACS RNA samples they will use existing lab Medicago single cell type gene expression data with the aim of identifying genes that control nodulation, mediate nitrogen and symbiont recognition responses and identify miRs that are predicted to control these genes as putative master regulators. These miRs will then be profiled in the same FACS RNA samples to test the predictions and develop miR-gene regulatory modules. The TaqMan kits allow us to accurately assay individual isoforms of miRs in the small amounts of total RNA generated from individual cell types. We could also use RNAseq for global miR level analysis but the value of predicting specific miRs to test from our existing FACS data means that we can drill down on the most important miRs straight away. For miRs that we have expression evidence for we will create miR perturbation lines to analyse the functional and phenotypic effects, predicting altered nodulation or lateral root development phenotypes. This work will involve developing your basic molecular biology and genetics skills and techniques in tissue culture and physiological investigation. To understand whether the miRs act to control conserved developmental processes we will make use of our parallel timeseries data for nitrogen and Rhizobium responses in the non-nodulating Arabidopsis within the prediction process. What theoretical methods will be developed and/or used? A range of bioinformatic techniques will be used to identify groups of nodulation and lateral root development-controlling genes from the gene expression data in both Medicago and Arabidopsis. The genes will then be analysed to identify miR recognition sites and the controlling miRs. To compare the sets of genes and controlling miRs between species the student will use existing methods to find orthologous genes between the species studied. Where we have sequenced genomes available we can make use of the gene position information to improve the quality of orthology assignments. Given expression data for different species, different cell types, and different time points we will identify groups of genes that share their temporal and spatial expression pattern across species. We will hypothesise that such sets of genes are regulated by a common miR regulatory mechanism. Why is this Systems Biology? One cycle of systems biology will be identifying regulated genes, predicting controlling miRs and then analysing the expression of the miRs to see if they fit the predictions. The second cycle revolves around the hypothesis that orthologous miRs will be regulated in similar ways during nodulation/lateral root responses across different species, again to be tested using miR assays. The final cycle postulates that if the miR regulates nodulation/lateral root development in Medicago, that perturbing it will affect that process phenotypically, and furthermore that it will affect parallel or similar developmental processes across legumes and non-legumes.