Supplementary Figures - Proceedings of the Royal Society B
advertisement
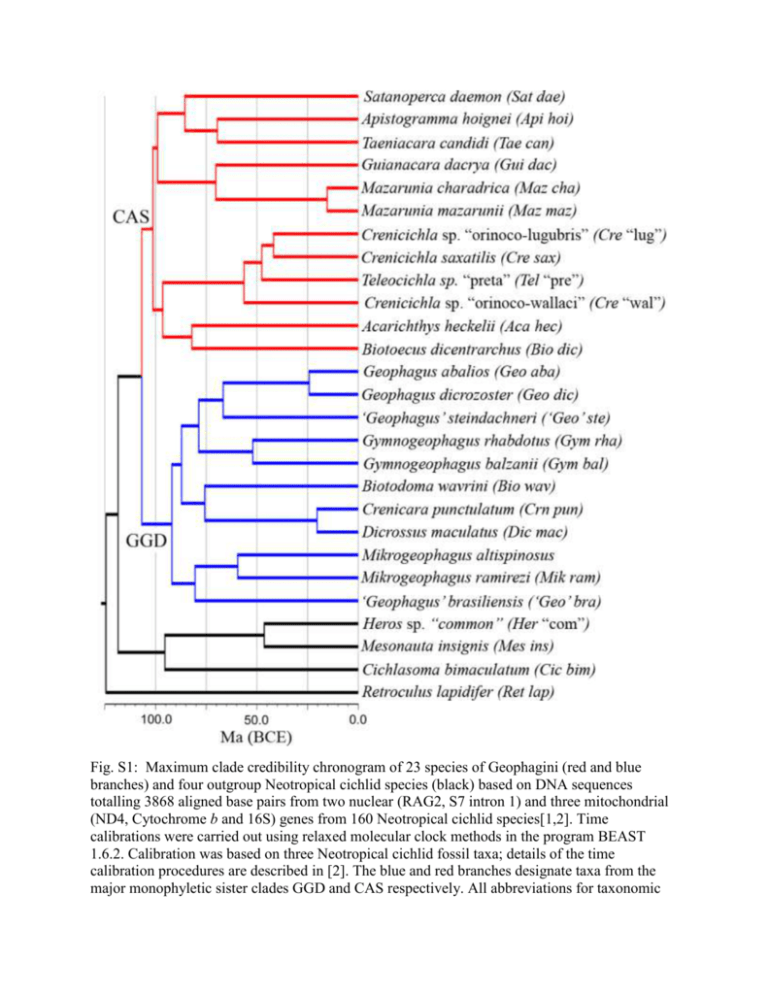
Fig. S1: Maximum clade credibility chronogram of 23 species of Geophagini (red and blue branches) and four outgroup Neotropical cichlid species (black) based on DNA sequences totalling 3868 aligned base pairs from two nuclear (RAG2, S7 intron 1) and three mitochondrial (ND4, Cytochrome b and 16S) genes from 160 Neotropical cichlid species[1,2]. Time calibrations were carried out using relaxed molecular clock methods in the program BEAST 1.6.2. Calibration was based on three Neotropical cichlid fossil taxa; details of the time calibration procedures are described in [2]. The blue and red branches designate taxa from the major monophyletic sister clades GGD and CAS respectively. All abbreviations for taxonomic names are given in Table S1. Most of the 27 taxa included in our analyses are the same as those used in previous phylogenetic analyses [1,2]. One exception to this is the inclusion of Crenicichla saxatilis in place of the closely related C. sveni (present in the previous 2010 phylogeny), another member of the C. saxatilis species group [3]. Fig. S2: Bite occlusion patterns in 8 species of Neotropical cichlids with their associated quadrate offset value. The angle of jaw occlusion increases steadily with increasing quadrate offset. Taxa illustrated from left to right are: (top row) Crenicichla sp. “Orinocowallaci”, Acaronia nassa, Petenia splendida, ‘Geophagus’ steindachneri, (bottom row) Andinoacara rivulatus, Uaru amphicanthoides, Symphysodon aequifasciatus and Herotilapia mutlispinosa. Fig. S3: A 3D functional phylomorphospace illustrating the relative lineage density and morphometric branch lengths of the two major clades of Geophagini. Points indicate the PC scores across the three critical axes of variation, and lines illustrate phylogenetic relationships between taxa. Point size and colour intensity illustrate depth along the horizontal plane of the image along which PC1 and PC2 vary (e.g. lighter objects are further into the background), while PC3 is oriented with the vertical plane of the image. A) Phylomorphospace of clade GGD (denser, comparably shorter morphometric branch lengths). B) Phylomorphospace of clade CAS (less dense, comparably longer morphometric branch lengths). Literature Cited 1 López-Fernández, H., Winemiller, K. O. & Honeycutt, R. L. 2010 Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae). Molecular phylogenetics and evolution 55: 1070–86. 2 López-Fernández, H., Arbour, J. H., Winemiller, K. O. & Honeycutt, R. L. 2013. Testing for ancient adaptive radiations in Neotropical cichlid fishes. Evolution 67: 1321-1337. 3 Kullander, S., Noren, M., Frioriksson, G. B. & Santos de Lucena, C. A. 2009 Phylogenetic relationships of species of Crenicichla (Teleostei: Cichlidae) from southern South America based on the mitochondrial cytochrome b gene. Journal of Zoological Systematics and Evolutionary Research 48: 248–258.