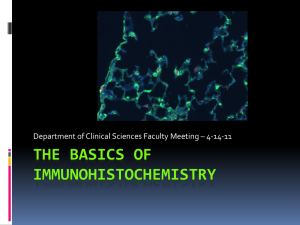
Fluorescence microscopy lab 2013
advertisement

Fluorescence Microscopy Lab Mark Berryman MCB 730 04/08/13 Introduction The utilization of fluorescent probes for microscopy has exploded over the past two decades. Specimen analyses range from examination of subcellular architecture to the study of whole organisms using both fixed and living cells and tissues. In addition to vital dyes and fluorescent-labeled proteins such as those commonly tagged with GFP (green fluorescence protein), the detection of proteins using indirect immunofluorescence microscopy is routine in contemporary cell and molecular biology research. Specimens can be stained and examined while alive or after preservation with chemical fixatives. Typically, visualization of the specimen is accomplished through the use of an epifluorescence microscope or a scanning confocal microscope: images can be recorded electronically or on film. Video tutorial on fluorescence http://probes.invitrogen.com/resources/education/ Fluorescence techniques http://probes.invitrogen.com/handbook/sections/0001.html More tips on techniques http://swehsc.pharmacy.arizona.edu/micro Compendium of fluorescent probes via http://probes.invitrogen.com/gallery/theartofimaging/ Nobel Prize in Chemistry 2008 http://nobelprize.org/nobel_prizes/nobelguide_che.pdf Overview of Today’s Exercise Cultured cells grown on glass coverslips will be fixed, stained, examined, and photographed. Your assignment is to compare the effects of two different fixation methods, formaldehyde versus methanol, on the ability to visualize cytoskeletal filaments in cultured mammalian cells. You will work in pairs: one partner will stain cells with a monoclonal antibody against ezrin (an actin-associated protein) and fluorescent phalloidin (a probe that binds to actin filaments) and the other partner will stain cells with a monoclonal antibody against beta tubulin and phalloidin. You will first examine the specimens using conventional epifluorescence microscopy and compare the effects of the two fixation conditions on the antibody staining and the phalloidin staining. Next, you will document the results using a digital camera. Finally, you will use the confocal microscope to record photographs of the fixation method that gives the best results. Fixation Methods: These are generic fixation procedures, although it should be noted that either formaldehyde or methanol can destroy many antigens. Therefore, it is wise to always compare at least two different fixation conditions for each antibody studied, ideally a crosslinking fixative (e.g. formaldehyde) versus a precipitating fixative (e.g methanol). I. Formaldehyde fixation 1) Fix cells in electron microscopy grade 4% formaldehyde/PBS for 8 min at room temperature. 2) Rinse cells several times in PBS. 3) Permeabilize cells in 0.1% Triton X-100/PBS for 60 sec. 4) Rinse cells briefly in PBS. Rinse again in fresh PBS. I. Methanol fixation 1) Immerse cells in ice-cold (-20C) absolute methanol containing 10mM EGTA for 2 minutes. 2) Rehydrate cells in PBS. Fluorescence Microscopy Lab Mark Berryman MCB 730 04/08/13 The cells have already been fixed and permeabilized. The cells are LLC-PK1 CL4 epithelial cells derived from pig kidney. In general, the cells will be triple-labeled, so stained structures will be red (alexa-546), green (alexa-488), and blue (DAPI). The following antibodies/fluorescent probes should stain the following organelles/structures: Mouse monoclonal E7 anti-β-tubulin (DSHB): microtubules. Use Goat anti-mouse IgG Alexa-488 secondary (green fluorescence) antibody. Mouse monoclonal CPTC-EZRIN anti-ezrin (DSHB): marker for cortical actin cytoskeleton, especially concentrated in microvilli. Use Goat anti-mouse IgG Alexa-488 secondary (green fluorescence) antibody. Phalloidin: a mushroom toxin that binds avidly to F-actin. The toxin is directly labeled with Alexa-546 (red fluorescence). DAPI: nucleic acids. The DAPI fluoresces blue when illuminated with ultraviolet light. Choose one antibody combination below. Primary Antibody E7 Tubulin mAb CPTC-EZRIN mAb 5) 6) 7) 8) 9) 10) 11) 12) Secondary Antibody/fluorescent probes GAM-Alexa 488 & phalloidin-Alexa 546 GAM-Alexa 488 & phalloidin-Alexa 546 Cell type LLC-PK1 Incubate cells in primary antibody (20μl) diluted in PBS for 30 min: place coverslip in humid chamber at room temperature. Rinse cells in PBS for 5 min: dip coverslips several times. Incubate cells in appropriate secondary antibody containing phalloidin diluted in PBS for 30 min. Rinse cells in fresh PBS for 5 min. Mount coverslip on a clean glass slide: quickly drain excess PBS on a piece of filter paper, then gently place coverslip on a small amount (6.5μl) of commercial antifade mounting medium (ProLong Gold: Molecular Probes). The mounting medium contains DAPI, a dye that stains DNA Place slide in dark overnight at room temperature to allow mounting medium to cure. Carefully seal edge of coverslip with clear nail polish. Allow polish to dry. Rinse the surface of the coverslip with water to remove salt. Blot dry with edge of kimwipe—do not touch coverslip. Store slide at -20°C. General Notes: (a) The concentration of fixative and duration of fixation should be optimized empirically for each antigen and antibody studied. (b) Cells can also be fixed and simultaneously permeabilized using organic solvents such as absolute ethanol or acetone and then rehydrated in PBS. Note that methanol fixation precludes visualization of the actin cytoskeleton with fluorescent-labeled phalloidin. Various methods of permeabilization should be compared to test for retention of antigenicity and possible alterations in antigen distribution that may result from extraction of cellular membranes. (c) Background staining may be minimized by the inclusion of a suitable blocking protein (e.g. 1-5% normal goat serum) or a low concentration of detergent (e.g. 0.05% Tween 20) during antibody incubations. (d) Never allow cells to dry during staining or mounting. Also, be careful not to move or touch the coverslip when applying nail polish. Fluorescence Microscopy Lab Mark Berryman MCB 730 04/08/13 Assignment 1. After preparing your specimens, you will observe them using epifluorescence microscopy on Wednesday. Important lessons in this assignment are: a. Careful observations and handwritten notes on what you observe while looking through the microscope. Do you see a convincing signal with each of the fluorescent stains? Where do the probes localize within the cell? Is there variability in localization or staining intensities among the cells? Do cells in mitosis offer any assistance in evaluating the results? b. Comparing fixation conditions. How did formaldehyde versus methanol affect staining intensity, subcellular localization, or consistency of staining? Which worked the best for the antibody you used? Which worked the best for the phalloidin? Do you see any differences in the DAPI staining pattern relating to the fixation method? c. Digital photography using epifluorescence. You should use the appropriate objective lens (magnification) to demonstrate a point in your specimen. Also, the exposure time should be appropriate so as not to under- or over-represent the staining in the real image you see under the microscope. For each fixation condition, you should show a representative field of each of the three channels at the same focal plane as well as a merged image of all three to show the spatial relationship between the stains. Prepare and label a figure as if presented in a journal article (e.g. Journal of Cell Biology or Molecular Biology of the Cell) along with a legend to describe what is shown in the image. Also, indicate the magnification used to capture your image. d. Confocal imaging. Using the best fixation for your antibody, scan a representative filed at a magnification similar to the one you used for the epifluorescence microscope images. Be sure to include a scale bar in the final image and record the thickness of the optical section. How do the images taken with the epifluorescence differ from those on the confocal? What are some advantages and disadvantages of scanning confocal microscopy? Please bring a blank CD or other electronic storage medium to store your data. 2. Assemble your best photographs into a single color figure as if it were being submitted for publication. Individual panels should be clearly labeled. Include a scale bar. Provide an accompanying figure legend (one paragraph) that concisely describes the specimen (i.e. cell type), method of fixation, and the reagents used for staining. Briefly describe the subcellular distribution of the proteins you stained in terms of their known functions. Incorporate sufficient background information about the proteins being localized and the pattern of stained structures so that a naïve reader can understand your description of the salient features of the figure (e.g. stage of cell cycle, subcellular distribution of protein, structures with prominent staining, spatial-temporal relationship between stained structures, etc…). 3. On a separate page, describe at least one experiment using fluorescence microscopy that would enhance your thesis/dissertation project. Be sure to include a description of any controls to account for antibody specificity and background fluorescence staining. 4. Please submit your assignment by email (berryman@ohiou.edu) no later than April 23. Score will be reduced by one letter grade for each day past the deadline.