mec12636-sup-0001-SupplementMaterial
advertisement

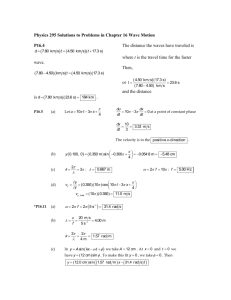
Bombus RAD tag Diversity Supplemental Material J. Lozier Supplementary Material Table S1 Sample locations for individuals used for RAD tag sequencing Species County State Latitude Longitude Sequence Reads (106) B. impatiens Bibb Alabama 33.05 -87.01 2.57 Washington Arkansas 35.82 -94.16 5.11 Hartford Connecticut 41.77 -72.90 1.74 Crawford Iowa 41.99 -95.39 4.00 Ogle Illinois 41.98 -89.36 4.10 Peoria Illinois 40.83 -89.80 7.42 Montgomery Indiana 39.96 -87.07 3.00 Ripley Indiana 39.07 -85.44 6.63 Ossage Kansas 38.64 -95.60 3.31 McCracken Kentucky 37.03 -88.76 3.08 Franklin Kentucky 38.16 -84.94 3.24 Winona Minnesota 43.98 -91.43 6.13 1 Bombus RAD tag Diversity Supplemental Material B. pensylvanicus J. Lozier Franklin Missouri 38.48 -90.82 16.6 Stokes North Carolina 36.47 -80.39 1.58 Seneca New York 42.68 -76.85 4.31 Belmont Ohio 40.00 -81.14 4.73 Cameron Pennsylvania 41.40 -78.03 3.80 Kershaw South Carolina 34.16 -80.57 1.33 Cocke Tennessee 35.92 -82.98 5.09 Appomattox Virginia 37.26 -78.68 1.13 Windsor Vermont 43.41 -72.71 3.62 Dane Wisconsin 43.04 -89.43 3.17 Bibb Alabama 33.05 -87.01 2.40 Prowers Colorado 38.11 -102.31 7.65 Columbia Florida 29.89 -82.67 7.02 Crawford Iowa 41.99 -95.39 2.95 Union Illinois 37.45 -89.12 8.27 2 Bombus RAD tag Diversity Supplemental Material J. Lozier Piatt Illinois 40.01 -88.64 7.88 Cherokee Kansas 37.34 -94.83 5.23 Saline Kansas 38.70 -97.43 6.80 Bossier Louisiana 32.53 -93.68 3.32 Lawrence Missouri 36.96 -93.68 7.24 Boone Missouri 38.99 -92.52 4.17 Franklin Missouri 38.48 -90.82 7.27 Clay Mississippi 33.55 -88.64 8.02 Lenoir North Carolina 35.29 -77.79 6.90 Howard Nebraska 41.13 -98.55 1.72 Wayne Ohio 40.91 -81.98 5.61 Cleveland Oklahoma 35.25 -97.27 6.72 Marion South Carolina 34.23 -79.15 4.47 Jasper South Carolina 32.59 -81.21 5.88 Stanley South Dakota 44.29 -100.33 2.41 3 Bombus RAD tag Diversity Supplemental Material J. Lozier Bastrop Texas 30.35 -97.38 7.69 Eastland Texas 32.10 -98.96 5.08 Greensville Virginia 36.64 -77.56 3.83 4 Bombus RAD tag Diversity Supplemental Material J. Lozier Impacts of read depth variation B. pensylvanicus RAD tags received, on average, higher sequencing coverage than B. impatiens RAD tags (in part due to the greater number of RAD tags in B. impatiens). Although sequencing depth in both species is very high (average >100x reads per site per bee; Fig. S1), which should facilitate accurate calling of heterozygous sites, I aimed to examine how this difference might bias diversity estimates. I filtered the SNP set to only consider those SNPs that received 125200X read depth, a region of substantial coverage overlap for the two species (Fig S1). I focused on comparing data sets for Filter Set 4 (20 individuals per species with the highest sequencing coverage; maximum of 5% missing data per SNP) to avoid sample size differences. Overall diversity estimates were slightly reduced in both species by this standardization, but SNP (heterozygosity per SNP) was similar for the two species, and RAD (average nucleotide diversity per entire RAD tag) was slightly higher in B. pensylvanicus, as in the full data set (Table S1). The site frequency spectra are also highly similar for the two data sets, with a slight increase in the proportions of low-frequency alleles in both species (Fig S2), explaining the slightly lower nucleotide diversity estimates. Thus, the relative comparison is not altered by limiting the analysis to SNPs with similar coverage. 5 Bombus RAD tag Diversity Supplemental Material J. Lozier Fig. S1 Average sequencing coverage per single nucleotide polymorphism per individual (a) and number of single nucleotide polymorphisms per RAD tag locus (b) following initial filtering (filter set 1). Invariant RAD tag loci not shown. 6 Bombus RAD tag Diversity Supplemental Material J. Lozier Table S2 Parameters estimated by filtering Filter Set 4 to mean SNP coverage of 125-200X per SNP per individual Mean read depth per SNP SNP No. No SNPs per Ind SNP SE RADtags RAD RAD SE B imp 3029 158.508 0.133 0.002 1932 0.0023 0.000055 B pen 2106 165.093 0.136 0.003 1211 0.0026 0.000078 0.7 impfil4 Proportion of SNPs 0.6 penfil4 0.5 impfil4_125-200x 0.4 penfil4_125-200x 0.3 0.2 0.1 0. 0.05 0.1 0.15 0.2 0.25 0.3 0.35 0.4 0.45 0.5 Minor Allele Frequency Figure S2 SNP minor allele frequency histograms with the “125-200X per site per individual read depth” filter [shows both the original ‘filter set 4’ data (solid colors), and for filter set 4 using the depth cutoff of 125-200x (solid+hatch)]. 7 Bombus RAD tag Diversity Supplemental Material J. Lozier Simulations of Diversity Under Population Declines Coalescent simulations were conducted in SIMCOAL 2.1.2. A historical population size of 50,000 haplodiploid individuals (= 75,000 chromosomes) was assumed, which provides a per site = 0.0015 with = 1 x 10-8 per site per generation in a stable population, similar to that seen in the present sequence data (see Results). A microsatellite = 5 x 10-5 per generation was applied, selected to produce a mean gene diversity of ~0.75 for a stable population. A series of instantaneous bottleneck sizes was considered starting 75 generations before present. 100 replicates of either ten microsatellite loci or 10,000 90 bp DNA sequence loci (no intralocus recombination, 3:1 Ts/Tv ratio) were simulated for each scenario and mean microsatellite heterozygosity or per-site nucleotide diversity for each replicate were analyzed in ARLEQUIN (references in main text). Contemporary eastern Bombus populations are essentially unstructured, with near-zero FST, D, no clustering in Bayesian approaches like STRUCTURE, and essentially no isolation by distance. For an unstructured population with starting diversity similar to B. impatiens (stable), results show that massive bottlenecks to 50-500 individuals would be needed to reduce microsatellite heterozygosity by ~20%, the proportional difference currently observed between B. pensylvanicus and B. impatiens whether individual populations are examined, or a pool of single individuals from separate populations (see main text). Per site nucleotide diversities are smaller than multilocus heterozygosity estimates and thus the absolute difference in diversity is smaller, but the proportional loss in diversity is similar for RAD tag-like data (Fig. S3). Also note that the RAD-tag like data had less variability among replicates, with no overlap in confidence regions among scenarios, as expected given the far greater number of loci (Fig. S3). 8 Bombus RAD tag Diversity Supplemental Material J. Lozier Thus RAD-tags should be capable of resolving differences in diversity due to demographic effects of sufficient impact to alter microsatellite gene diversity. Table S3 Coalescent simulation parameter summary Locus Type Demography Ancestral Ne (haplodiploids) Current Ne Msat Msat Msat Msat 90bp sequence (RAD-like) 90bp sequence (RAD-like) 90bp sequence (RAD-like) Stable Bneck-75 gen bp Bneck-75 gen bp Bneck-75 gen bp Stable 50,000 50,000 50,000 5,000 Sample Size (diploids) 20 20 Independent Loci Mutation Rate 10 10 5.00E-05 5.00E-05 50,000 500 20 10 5.00E-05 50,000 50 20 10 5.00E-05 50,000 50,000 20 10,000 1.00E-08 Bneck-75 gen bp Bneck-75 gen bp 50,000 500 20 10,000 1.00E-08 50,000 50 20 10,000 1.00E-08 Figure S3 Box plots of mean diversity from 100 simulation replicates per scenario for microsatellite (left) and RADtag-like (right) data under a range of bottlenecks from an ancestral Ne = 50,000 haplodiploids. See Table S3 for simulation parameters. Note that based on microsatellite simulation results, the 5,000 Ne bottleneck was not performed for the more computationally intensive RADtag-like data as differences from the stable population simulation should be small. 9 Bombus RAD tag Diversity Supplemental Material J. Lozier Quality checks to ensure integrity of sample data One concern with RAD tag and other barcoded NGS library approaches is the potential for laboratory or computational errors to result in erroneous data sets. Such errors can be difficult to detect because of the massive amounts of data produced by these methods prohibit visual screening of samples for inconsistencies. Of particular interest in the present study is the potential for barcode-swaps during library preparation or demultiplexing to result in mixed data sets, which would corrupt any between species comparisons. I took advantage of the evolutionary distance between B. impatiens and B. pensylvanicus to check the integrity of sequence data within each species. For this analysis, I took the two data sets reported in the study (B. imp aligned against the B. imp RAD reference and B. pen aligned against the B. pen RAD reference) and made use of alignments for both B. impatiens and B. pensylvanicus against a common B. impatiens reference (not otherwise discussed in the main paper) using Bowtie and filtered as for Filter Set 1 (see Methods). The latter analysis produces data of a characteristic form: a large and nearly identical fraction (~45%) of the called SNPs are fixed for the alternate allele state across all B. pensylvanicus individuals, as one might expect for such deeply divergent lineages (Figure S4), while no B. impatiens individuals show this pattern. Propor on of Sites Fixed for non B. impa ens reference allele (alt allele) 0.5 0.45 0.4 0.35 0.3 0.25 0.2 impa ens pensylvanicus 0.15 0.1 0.05 0 Figure S4 Proportion of fixed non-B. impatiens-reference alleles when reads from individuals in both species are simultaneously aligned against a B. impatiens reference. Each point = 1 individual. 10 Bombus RAD tag Diversity Supplemental Material J. Lozier A PERL script by C. Bergey (https://code.google.com/p/vcf-tab-to-fasta/) was then used to convert the vcf files to fasta format (missing data = -, heterozygotes = IUPAC ambiguities) as a second approach to detect outlier individuals. MEGA 5.2.2 (Tamura et al., 2011) was used to construct neighbor-joining trees, deleting missing data and calculating branch lengths as the number of differences. Figure S5 shows the resulting trees. As expected for very-low FST species, the data sets used in the main paper show unstructured star-like trees with no geographical patterns apparent and small numbers of differences separating individuals. The lower tree shows an analysis with both species aligned to the B. impatiens reference and shows a large divergence of several thousand fixed base differences between the two species, and no individuals sorting incorrectly among species. Such patterns suggest no major errors in read identity across samples consistent with barcode swaps. Subsequent errors in post-alignment processing of BAM files could be detected by comparing VCF SNP call files with those produced by independent analysis of the original alignment files with ANGSD. Overall, similar sites were called as variable in both sets, and comparison of allele frequencies in the two analyses show comparable nucleotide diversities (main text). Differences appear mostly due to the larger number of regions including in the ANGSD analysis, and a minor differences for sites that were filtered out of the VCF-formatted genotyping data (e.g., due to low coverage, which ANGSD allows and takes into account), or due to the fact that ANGSD is estimating nucleotide diversities by taking into account multiple factors (sequence depth, quality, etc), and not calculating them solely from allele counts. These checks suggest that nothing erroneous occurred during post-alignment processing of bam files. The vcf files for the two species, the per site ANGSD diversity output, and original alignments, are located on DRYAD. Based on these data checks, incorrect pooling of data among samples 11 Bombus RAD tag Diversity Supplemental Material J. Lozier and species would also presumably result in larger numbers of SNPs per RAD tag, as well as elevated intermediate allele frequencies in the site frequency spectra, rather than the typical “Lshaped” distributions observed here. Together, these analyses suggest that the present RAD tag data contain no major contamination errors that would affect inferences. Figure S5 NJ trees for RAD tag reads aligned to within species (top) and for both species together aligned against the B. impatiens reference (bottom). Scale provided as number of differences. 12 Bombus RAD tag Diversity Supplemental Material J. Lozier Tamura K, Peterson D, Peterson N et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–9. 13