click here
advertisement

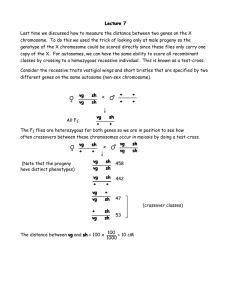
1. In birds, the females are the heterogametic sex. ZcZc (male) x ZB+W (female) ZcZB+ (male) and ZcW females Ans. (c) blue males and cameo females 2. To produce 3 distinct phenotypes, the parents have to be heterozygous for all three alleles, given this dominance relationship: MR Md Md MR Md M dM d M MR M MM d Ans: (c) MRMd x MMd 3. In this cross, you have to remember that the Yellow allele is a recessive lethal, and that YY individuals are never born. Individuals with a yellow phenotype are therefore obligate heterozygotes, and the ratio of yellow:agouti will be 2/3 Yy and 1/3 yy in any cross between two yellow individuals. The fuzzy allele (s) is recessive to the straight allele (S). The progeny are therefore the result of a dihybrid cross: YySs x YySs ¾ S_ 6/12 YyS_ (1/2 yellow, straight) 2/3 Yy ¼ ss 2/12 Yyss (1/6 yellow, fuzzy) ¾ S_ 3/12 yySs (1/4 agouti, straight) ¼ ss 1/12 yyss (1/12 agouti, fuzzy) 1/3 yy Ans: (d) 2 and 4 are correct. 4. The first cross will create heterozygous females; when crossed to hornless males: males ch c+ c + c + c h c + c + 1/2 of males c + c+c h c +c + females c h c + c + c + c h c + c + no females c + c+c h c +c + Ans: (d); ½ males, no females. The parental classes are the largest (+r+/b+s); to establish which gene is in the middle, you need to examine what gene is exchanged to generate the smallest class, the double recombinant, since a crossover here has occurred on either side of the middle gene. In this case, it is the b locus that changes relative to the parentals (br+/++s), so the starting alignment and phasing relationships in the heterozygote parent are: r + + + b s phenotype +rs br+ +++ +r+ brs ++s b+s b++ number of progeny 42 2 19 189 13 1 204 30 The map distance between the r and b loci is calculated as: Single crossovers: rbs 13 (single crossover) +++ 19(single crossover) rb+ 2 (double crossover) ++s 1 (double crossover) 35/500 = 0.07 or 7 map units The map distance between the r and s loci is calculated as: Single crossovers: r+s 42 (single crossover) +b+ 30 (single crossover) rb+ 2 (double crossover) ++s 1 (double crossover) 75/500 = 0.15 or 15 map units 5. The distance between the r and s loci is 7 + 15 or 22 map units. (d). The expected number of DCOs for this experiment, assuming no interference, is: (0.07)(0.15)(500) = 5.25 double crossovers The coefficient of coincidence is 3/5.25 = 0.57 Interference is therefore 1- coc = 1- 0.57 = 0.43 6. Ans: 0.43 (c). Looking at this data set, we do not see 4 distinct classes of values- there are really just two sets. This suggests that one gene pair is linked, but the other is independently segregating from the other two. If this is the case, we expect it to will show independent assortment. Are the l and g genes linked? lg 20 +110 = 130 ++ 102 + 22 = 124 l+ 106 + 23 = 129 +g 18 + 99 = 117 essentially a 1:1:1:1 ratio; these two genes look like they are assorting independently. 7. The genes show no evidence of linkage (e). Are the t and g genes linked? tg 99 + 110 = 200 ++ 102 + 106 = 208 +g 18 + 20 = 38 t+ 22 + 23 = 45 These genes look linked, and in the following phasing relationship: t g + + 8. The distance between the t and g genes is : (38+45)/500 = 0.166 or 16.6 map units (c). 1 ab ab ab ab ++ ++ ++ ++ 30 2 ab ab a+ a+ +b +b ++ ++ 8 3 ab ab +b +b ++ ++ a+ a+ 5 Tetrad Classes 4 5 a+ a+ a+ a+ a+ ab a+ ab +b +b +b +b +b ++ +b ++ 30 10 6 +b +b ab ab a+ a+ ++ ++ 7 7 +b +b ++ ++ ab ab a+ a+ 8 8 a+ a+ +b +b ab ab ++ ++ 2 9. ½ (5 + 7 + 2)/100 = 0.07 = 7 m.u. between centromere and a locus. Ans. (c). 10. PD ≈ NPD (classes 1 and 4); the a and b gene loci are not linked. ANS. (e).