Teaching Notes
advertisement

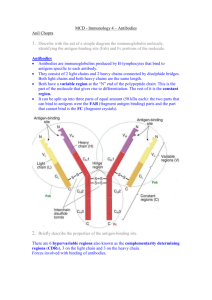
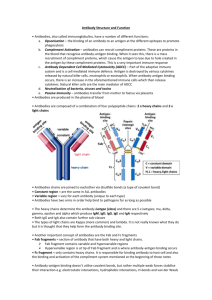
Exploring a Protein Structure in the RCSB PDB: Antibodies Level: Advanced Teaching Notes Learning Goals: 1. Visualize the structure of a given molecule using RCSB PDB resources. 2. Explore the structure to understand its structure function relationships Educational Standards A. Common Core a. Craft and Structure i. RI.9-10.4 ii. RI.11-12.4 b. Integration of Knowledge and Ideas i. RI.9-10.7 ii. RI.11-12.7 B. Next Generation Science Standards a. Practices i. 8. Obtaining, Evaluating and Communicating Information b. Crosscutting Concepts i. 3. Scale, proportion and quantity ii. 4. Systems and system models iii. 6. Structure and function c. Disciplinary Core Ideas i. LS1.A: Structure and Function ii. PS2.B: Types of Interactions C. Advanced Placement Biology - Essential Knowledge (EK), Learning Objectives (LO), Science Practices (SP) a. EK 4.A.1 i. LO 4.2, SP 1.3 ii. LO 4.3, SP 6.1, 6.4 Teaching Notes: About Antibodies: 1. Structure: a. The antibody IgG is composed of 4 protein polymer chains – two light and two heavy chains. b. Each of the light chains has two beta-sandwich domains (called immunoglobulin domains), while each heavy chain has four such domains. c. The immunoglobulin domain has two beta sheets composed of 3-4 and 4-5 strands. d. A single disulfide bond stabilizes each of the beta sandwich structures. e. Additional disulfide bonds hold the 4 polymer chains together. f. A number of saccharides (sugars) are covalently linked to the heavy chains in the IgG stem (part of antibody not binding to antigens). This makes IgG a glycoprotein and imparts structural stability to the molecule. 2. Function: Developed as part of the RCSB Collaborative Curriculum Development Program 2015 a. b. c. d. Level: Advanced Teaching Notes The overall structure of the antibody is “Y-shaped” and has two antigen binding sites. The immunoglobulin domains closest to the antigen binding regions vary from antibody to antibody resulting in specific antigen-antibody binding. In the variable domain there are specific loops that directly touch the antigen and bind to it. The sequences of these loops are highly variable, imparting specificity to the antigen-antibody interactions. Each antigen-binding site is composed of three hyper-variable loops from the heavy chain and three from the light chain. Activity Suggestions: 1. You may want your students to attempt this activity before they make the antibody paper model so that they are oriented to the overall shape and structure of the antibody IgG. 2. Review the IgG structure with your students to make sure that they notice all the structural details described above. Answers to Questions in Exercise: Q1. What is the predominant secondary structural element seen in the immunoglobulin structure? A1: The immunoglobulin domains have beta sheets, thus the predominant secondary structure seen in the molecule is beta-sheet. Q2. What do you think the non-standard residues are? What is their function? A2: These are saccharide residues (sugars), glycosylating the Ig heavy chains. These residues impart structural stability to the stem region of the IgG molecule. Q3. Draw a picture of the immunoglobulin molecule and label the heavy and light chains. Save a suitable image and label it. Also label the antigen binding sites. A3: Fab region Light chain Light chain AnAntigen gen binding site site binding An gen binding site Variable region Fc region Heavy chain Papain cleavage sites Variable region Constant region Heavy chain PDB IDs 1igt, 3hfm Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Level: Advanced Teaching Notes Q4. Where are these (Cys) residues located? Can you explain the role that these residues play in the stability of the antibody structure? A4: There are 2 Cys residues in each Ig domain that forms a disulfide bridge stabilizing the domain. In addition there are Cys residues in the random coiled regions of the Ig chains that form S-S bonds to covalently link the Ig chains to each other. Q5. How well do the 2 structures (PDB entry 1igt and 1igy) match? Do they align? in which parts? A5: These two structures superpose only in the stem (Fc) region of the IgG molecule. Arms of the antibody (with the antigen binding sites) are not aligned. Q6. What can you say about Antibody structures based on this comparison? (Hint: Are they rigid or flexible). A6: The structure of the Fc region is stable and rigid. This is probably required for it to be able to bind to Fc receptors on cells. While the structures of the immunoglobulin domains are stable, there is flexibility in binding to the antigens (especially at the hinge regions), allowing the antibody to bend and twist in order to bind to the antigens. An image of the superposed structures of PDB entry 1igt and 1igy is shown below: Developed as part of the RCSB Collaborative Curriculum Development Program 2015