mec12544-sup-0002-FigureLegends
advertisement

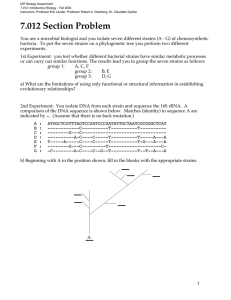
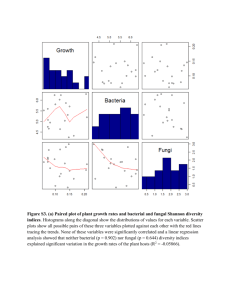
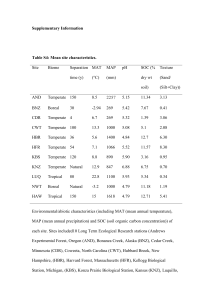
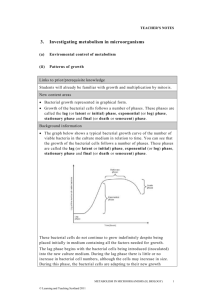
1 Supplementary Figure Legends 2 3 Fig. S1 Rarefaction curves for the amplicon pyrosequencing profiles of bulk soil samples from 4 Japanese barberry using genomic DNA that was either sequenced individually or pooled. Panels 5 (a) and (b) present the number of observed phylotypes as a function of the total number of 6 sequences for bacterial 16S rRNA fungal ITS assays, respectively. These data demonstrate 7 similar OTU richness in the individual (B2.1, B2.2, B2.3) and pooled (B2) genomic DNA 8 samples from the bulk soil. 9 10 Fig. S2 The bacterial community profiles obtained with 16S rRNA amplicon pyrosequencing of 11 the bulk soil genomic DNA samples when sequenced as a pooled sample (B2) versus 12 individually (B2.1, B2.2, B2.3). A weighted UniFrac significance test indicates there is no 13 significant difference between the bacterial communities observed in these samples (p > 0.05). 14 Colored bars represent the proportion of each taxa in the total bacterial community of the sample. 15 16 Fig. S3 The fungal community profiles obtained with ITS amplicon pyrosequencing of the bulk 17 soil genomic DNA samples when sequenced as a pooled sample (B2) versus individually (B2.1, 18 B2.2, B2.3). A weighted UniFrac significance test indicates there is no significant difference 19 between the bacterial communities observed in these samples (p > 0.05). Colored bars represent 20 the proportion of each taxa in the total bacterial community of the sample. 21 22 Fig. S4 Rarefaction curves charting the number of observed phylotypes as a result of the total 23 number of sequences obtained per sample. The bacterial (16S rRNA) and fungal (ITS) 24 rarefaction curves are shown in (a) and (b), respectively. These data indicate the current depth of 25 sequencing has not revealed the total microbial communities, although more of the fungal 26 community members were detected than the bacterial community members (as determined by the 27 slope of the curve). 28