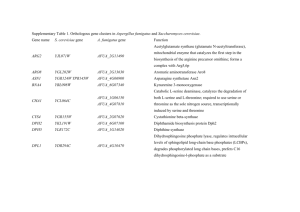
Table S2. Genes down-regulated by PUFAs Gene Name Description
advertisement

Table S2. Genes down-regulated by PUFAs Gene Description Name YJL179W bovine prefoldin subunit 1 homolog (putative) YOR244W Acetyltransferase in the SAS gene family YGL127C SOH1 encodes a novel 14-kD protein with limited sequence similarity to RNA polymerases. The Soh1 protein interacts with a DNA repair protein, Rad5p, in a two-hybrid system assay. YFR031C YKR024C YOR285W YDR144C YKL041W YER056C YLR011W YDR019C YPL045W YDR456W YMR004W YCR054C YBR034C YPR025C YDL100C YGR098C YOR076C YNL244C YML069W YKL122C YMR091C YHR010W YJL097W Fold Change 0,498 0,497 0,496 SMC chromosomal ATPase family member putative RNA helicase Protein of unknown function containing a rhodanese-like domain; localized to the mitochondrial outer membrane; protein abundance increases in response to DNA replication stress aspartyl protease related to Yap3p involved in secretion purine-cytosine permease FMN-dependent NAD(P)H:quinone reductase, may be involved in quinone detoxification; expression elevated at low temperature; sequesters the Cin5p transcription factor in the cytoplasm in complex with the proteasome under reducing conditions glycine cleavage T protein (T subunit of glycine decarboxylase complex Vacuolar sorting protein Na+\/H+ exchanger peripheral Golgi membrane protein CTR86 shares a terminator region with THR4. CTR86 contains aGCN4 responsive site suggesting it may also be involved in amino acid biosynthesis. 0,496 0,496 0,496 nuclear protein arginine methyltransferase (mono- and asymmetrically dimethylating enzyme) novel cyclin gene\; encodes subunits of TFIIK, a subcomplex of transcription factor TFIIH 0,492 Guanine nucleotide exchange factor for Gpa1p; amplifies G protein signaling; subunit of the GET complex, which is involved in Golgi to ER trafficking and insertion of proteins into the ER membrane; has low-level ATPase activity; protein abundance increases in response to DNA replication stress involved in regulation of spindle pole body duplication Coupling protein that mediates interactions between the Ski complex and the cytoplasmic exosome during 3'-5' RNA degradation; eRF3-like domain targets nonstop mRNA for degradation; null mutants have superkiller phenotype (2, 3, 4, 5, 6 and see Summary Paragraph) translation initiation factor 3 (eIF3) binds DNA polymerase delta component of signal recognition particle involved in nuclear protein targeting Ribosomal protein L27A Essential 3-hydroxyacyl-CoA dehydratase of the ER membrane, involved in elongation of very long-chain fatty acids; evolutionarily conserved, similar to mammalian PTPLA and PTPLB; involved in sphingolipid biosynthesis and protein trafficking 0,491 0,496 0,496 0,495 0,495 0,494 0,494 0,493 0,493 0,492 0,492 0,491 0,490 0,490 0,490 0,490 0,490 0,490 0,490 YLR126C YPL243W YNL250W YAL059W YDR043C YIL104C YDR212W YLR168C YHR103W YLL011W YLR408C YGL056C YDL120W YCR016W YOR252W YMR025W YPR101W YPL212C YMR074C YML017W YKL076C YML006C YBL004W YKL162C-A Putative glutamine amidotransferase; has Aft1p-binding motif in the promoter; may be involved in copper and iron homeostasis; YLR126C is not an essential protein; relocalizes from nucleus to cytoplasmic foci upon DNA replication stress component of signal recognition particle Contains a purine-binding domain, two heptad repeats and a hydrophobic tail. putative transmembrane domain protein involved in cell wall biogenesis transcriptional repressor which can bind to UAS-1 in the STA1 promoter and which can interact with Ssn6p 0,489 Chaperone protein required for the assembly of box H/ACA snoRNPs and thus for pre-rRNA processing, forms a complex with Naf1p and interacts with H/ACA snoRNP components Nhp2p and Cbf5p; homology with known Hsp90p cochaperones chaperonin subunit alpha possibly involved in intramitochondrial sorting involved in bud growth 56 kDa nucleolar snRNP protein that shows homology to beta subunits of Gproteins and the splicing factor Prp4 0,488 Putative protein of unknown function; likely member of BLOC complex involved in endosomal cargo sorting; green fluorescent protein (GFP)-fusion protein localizes to the endosome; YLR408C is not an essential gene similar to S. pombe sds23 mitochondrial protein that regulates mitochondrial iron accumulation Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; predicted to be involved in ribosome biogenesis Protein of unknown function that associates with ribosomes Subunit of the Cop9 signalosome, which is required for deneddylation, or removal of the ubiquitin-like protein Rub1p from Cdc53p (cullin); involved in adaptation to pheromone signaling; functional equivalent of canonical Csn6 subunit of the COP9 signalosome Component of a protein complex associated with the splicing factor Prp19p. intranuclear protein which exhibits a nucleotide-specific intron-dependent tRNA pseudouridine synthase activity 0,486 Protein with homology to human PDCD5; PDCD5 is involved in programmed cell death; N-terminal region forms a conserved triple-helix bundle structure; overexpression promotes H2O2-induced apoptosis; YMR074C is not an essential gene; protein abundance increases in response to DNA replication stress (putative) involved in mRNA splicing Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 69% of ORF overlaps the uncharacterized ORF YKL075C CAAX box containing protein Component of the small-subunit (SSU) processome, which is involved in the biogenesis of the 18S rRNA Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data 0,482 0,489 0,488 0,488 0,488 0,488 0,488 0,487 0,486 0,485 0,485 0,485 0,485 0,483 0,483 0,482 0,482 0,481 0,481 0,481 0,481 YNL227C YDL155W YOR220W YOR094W YDR080W YBR079C YCL067C YIL052C YNL001W YDR377W YER184C YDR503C YJL031C YGR057C YIL127C YIL138C YDR373W YPR108W YER073W YDR037W YPR119W YNL039W YPR046W YGL061C YJL190C YDL190C YLR056W YKL014C YBL059C-A YLR275W YDR082W YDR272W YLR210W YBR073W Co-chaperone that stimulates the ATPase activity of Ssa1p, required for a late step of ribosome biogenesis; associated with the cytosolic large ribosomal subunit; contains a J-domain; mutation causes defects in fluid-phase endocytosis G(sub)2-specific B-type cyclin Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; phosphorylated in response to alpha factor; protein abundance increases in response to DNA replication stress GTP-binding ADP-ribosylation factor component of vacuolar membrane protein complex translation initiation factor eIF3 Mating type protein alpha-2 Ribosomal protein L34B Involved in protein translation ATP synthase subunit f Putative zinc cluster protein; deletion confers sensitivity to Calcufluor white, and prevents growth on glycerol or lactate as sole carbon source Lipid phosphate phosphatase Geranylgeranyltransferase Type II alpha subunit (PGGTase-II, alpha subunit) involved in transport of nitrogen-regulated permeases Putative protein of unknown function; identified in a screen for mutants with decreased levels of rDNA transcription; green fluorescent protein (GFP)fusion protein localizes to the nucleolus; predicted to be involved in ribosome biogenesi Tropomyosin isoform 2 N-myristoylated calcium-binding protein that may have a role in intracellular signaling through its regulation of the phosphatidylinositol 4-kinase Pik1p; member of the recoverin/frequenin branch of the EF-hand superfamily Subunit of the regulatory particle of the proteasome mitochondrial Aldehyde Dehydrogenase lysyl-tRNA synthetase G(sub)2-specific B-type cyclin 90 kd subunit of TFIIIB, also called TFIIIB90 or B'' or B''90 component Involved in chromosome segregation Death Upon Overexpression Ribosomal protein S22A (S24A) (rp50) (YS22) ubiquitin fusion degradation protein C-5 sterol desaturase Nucleolar protein required for the normal accumulation of 25S and 5.8S rRNAs, associated with the 27SA2 pre-ribosomal particle; proposed to be involved in the biogenesis of the 60S ribosomal subunit Similar to very hypothetical S. pombe protein U1 snRNP protein of the Sm class involved in telomere length regulation Cytoplasmic glyoxylase-II G(sub)2-specific B-type cyclin Putative helicase similar to RAD54 0,480 0,480 0,479 0,479 0,478 0,478 0,478 0,478 0,478 0,478 0,478 0,478 0,476 0,476 0,476 0,476 0,476 0,476 0,475 0,475 0,475 0,475 0,474 0,474 0,473 0,473 0,472 0,472 0,470 0,469 0,469 0,469 0,468 0,468 YLR053C YCL056C Putative protein of unknown function Peroxisomal integral membrane protein that regulates peroxisome populations; interacts with Pex11p, Pex25p, and Pex27p to control both constitutive peroxisome division and peroxisome morphology and abundance during peroxisome proliferation Putative protein of unknown function RNA polymerase II subunit The TyB Gag-Pol protein. Gag processing produces capsid proteins. Pol is cleaved to produce protease, reverse transcriptase, and integrase activities. 0,468 0,467 YJR155W YGR160W Hypothetical aryl-alcohol dehydrogenase (AAD) Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data 0,464 0,462 YNL002C YGR246C Q0140 YDR323C Protein with similarity to ribosomal proteins including Rpl6p RNA polymerase III transcription factor with homology to TFIIB mitochondrial ribosomal protein encoded by mitochondrial genome. cytosolic and peripheral membrane protein with three zinc fingers\; cysteine rich regions of amino acids are essential for function 0,461 0,461 0,461 0,460 YOR078W YMR085W BUD site selection Putative protein of unknown function; YMR085W and adjacent ORF YMR084W are merged in related strains, and together are paralogous to glutamine-fructose-6-phosphate amidotransferase GFA1 kinesin-like protein Putative PINc domain nuclease required for early cleavages of 35S pre-rRNA and maturation of 18S rRNA; component of the SSU (small subunit) processome involved in 40S ribosomal subunit biogenesis; copurifies with Faf1p 18-kDa phosphotyrosine phosphatase of unknown function Subunit of the NuA4 histone acetyltransferase complex, which acetylates the N-terminal tails of histones H4 and H2A Ubiquitin; becomes conjugated to proteins, marking them for selective degradation via the ubiquitin-26S proteasome system; essential for the cellular stress response; encoded as a polyubiquitin precursor comprised of 5 head-to-tail repeats; protein abundance increases in response to DNA replication stress Ribosomal protein L26B (L33B) (YL33) Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene YPT6; deletion causes a vacuolar protein sorting defect Subunit of the regulatory particle of the proteasome involved in silencing at telomeres, HML, and HMR mitochondrial protein, prohibitin homolog\; homolog of mammalian BAP37 and S. cerevisiae Phb1p 0,460 0,460 Myristoylated subunit of ESCRTIII, the endosomal sorting complex required for transport of transmembrane proteins into the multivesicular body pathway to the lysosomal/vacuolar lumen; cytoplasmic protein recruited to endosomal membranes WD40 domain-containing protein involved in endosomal recycling; forms a complex with Rrt2p that functions in the retromer-mediated pathway for 0,456 YHR054C YGL070C YGR161WB YKL179C YDR339C YPR073C YNL136W YLL039C-R YGR034W YLR261C YDL097C YDR181C YGR231C YMR077C YPL183C 0,466 0,465 0,464 0,459 0,459 0,459 0,458 0,458 0,457 0,457 0,456 0,456 0,456 0,456 YDL133W YDR470C YGR272C YBR058C-A YOR189W YJL148W YPR048W YER050C YPR105C YKR031C YOR294W YOR327C YHR170W YKL173W YLR367W YOR305W YLR441C YBL003C YER099C YLR197W YBL090W YKL137W recycling internalized cell-surface proteins; evidence it interacts with Trm7p for 2'-O-methylation of N34 of substrate tRNAs; has a role in regulation of Ty1 transposition; human ortholog is WDR6 Regulator of phospholipase D (Spo14p); interacts with Spo14p and regulates its catalytic activity; capable of buffering the toxicity of C16:0 platelet activating factor, a lipid that accumulates intraneuronally in Alzheimer's patients Outer membrane component of the mitochondrial fusion machinery; binds directly to Fzo1p and Mgm1p and thus links these two GTPases during mitochondrial fusion; involved in fusion of both the outer and inner membranes; facilitates dimerization of Fzo1p during fusion; import into the outer membrane is mediated by Tom70p and Mim1p; has similarity to carrier proteins but is not likely to function as a transmembrane transporter Essential protein required for maturation of 18S rRNA; null mutant is sensitive to hydroxyurea and is delayed in recovering from alpha-factor arrest; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus involved in sphingolipid biosynthesis Component of the INO80 chromatiin remodeling complex and target of the Mec1p/Tel1p DNA damage signaling pathway; proposed to link chromatin remodeling to replication checkpoint responses unshared RNA polymerase I subunit Conserved NAPDH-dependent diflavin reductase; component of an early step in the cytosolic Fe-S protein assembly (CIA) machinery; transfers electrons from NADPH to the Fe-S cluster of Dre2p; plays a pro-death role under oxidative stress; Tah18p-dependent nitric oxide synthesis confers hightemperature stress tolerance protein of the small subunit of the mitochondrial ribosome Essential component of the conserved oligomeric Golgi complex (Cog1p through Cog8p), a cytosolic tethering complex that functions in protein trafficking to mediate fusion of transport vesicles to Golgi compartments Phospholipase D Regulator for ribosome synthesis vesicle-associated membrane protein (synaptobrevin) homolog cytoplasmic factor required for a late cytoplasmic assembly step of the 60S subunit U5 snRNP-specific protein related to EF-2 Ribosomal protein S22B (S24B) (rp50) (YS22) Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion; deletion confers sensitivity to 4-(N-(Sglutathionylacetyl)amino) phenylarsenoxide (GSAO); YOR305W is not an essential gene Ribosomal protein S1A (rp10A) Histone H2A (HTA1 and HTA2 code for nearly identical proteins) ribose-phosphate pyrophosphokinase 2 homology to microtubule binding proteins and to X90565_5.cds Component of the small subunit of mitochondrial ribosomes Evolutionarily conserved copper-binding protein of the mitochondrial intermembrane space, may be involved in delivering copper from the matrix to the cytochrome c oxidase complex; contains a twin CX9C motif 0,455 0,455 0,453 0,452 0,451 0,451 0,450 0,450 0,450 0,450 0,449 0,449 0,449 0,449 0,448 0,447 0,445 0,445 0,444 0,444 0,443 0,443 YHR001WA YER139C 8.5 kDa subunit of the ubiqunol-cytochrome c oxidoreductase complex 0,443 CTD phosphatase; dephosphorylates S5-P in the C-terminal domain of Rpo21p; has a cysteine-rich motif required for function and conserved in eukaryotes; shuttles between the nucleus and cytoplasm; RTR1 has a paralog, RTR2, that arose from the whole genome duplication high-temperature lethal 0,442 interacts with Nap1, which is involved in histone assembly Cytosolic Aldehyde Dehydrogenase Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed as polycistronic mRNA with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions YJR032W peptidyl-prolyl cis\/trans isomerase YLR392C Protein of unknown function that contains 2 PY motifs and is ubiquinated by Rsp5p; overexpression confers resistance to arsenite; green fluorescent protein (GFP)-fusion protein localizes it to the cytoplasm; non-essential gene YDR427W Subunit of the regulatory particle of the proteasome YPL222W Putative protein of unknown function; proposed to be involved in responding to environmental stresses; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies YJL074C SMC chromosomal ATPase family member YBL063W kinesin related protein YLR103C omosomal DNA replication initiation protein YJL208C mitochondrial nuclease YKR023W Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies YOR148C spliceosome-associated protein YGR150C Mitochondrial 15s rRNA-binding protein; required for intron removal of COB and COX1 pre-mRNAs; contains pentatricopeptide repeat (PPR) motifs; mutant is respiratory deficient and has defective plasma membrane electron transport YMR119W- Dubious open reading frame unlikely to encode a functional protein, based on A available experimental and comparative sequence data YML108W Protein of unknown function; structure defines a new subfamily of the split beta-alpha-beta sandwiches; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YML108W is not an essential gene; relative distribution to the nucleus increases upon DNA replication stress YIL035C alpha subunit of casein kinase II YMR299C Description YMR154C Cysteine protease similar to E. nidulans palB YNR032Cubiquitin-like modifier A YHR184W involved in meiosis, nuclear division and spore formation YOL080C RNA EXonuclease\; part of family of 3'-to5' exonucleases. See Moser et al. 1997 Nucleic acids Res. 25:5110-5118 0,440 0,440 0,438 YNR055C 0,426 YCR020WB YDR162C YPL061W YMR009W Putative ion transporter similar to the major facilitator superfamily of transporters 0,441 0,437 0,437 0,437 0,436 0,436 0,435 0,434 0,434 0,434 0,433 0,433 0,433 0,432 0,430 0,429 0,428 0,428 0,428 0,428 YHR055C YDR228C YGL164C copper-binding metallothionein Component of pre-mRNA cleavage and polyadenylation factor I RanGTP-binding protein, inhibits RanGAP1 (Rna1p)-mediated GTP hydrolysis of RanGTP (Gsp1p); shares similarity to proteins in other fungi but not in higher eukaryotes 0,426 0,425 0,424 YDL121C Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YDL121C is not an essential protein 40S ribosomal protein S27A (rp61) (YS20) Subunit of GID complex; involved in proteasome-dependent catabolite inactivation of gluconeogenic enzymes FBPase, PEPCK, and c-MDH; forms dimer with Rmd5p that is then recruited to GID Complex by Gid8p; contains a degenerate RING finger motif needed for GID complex ubiquitin ligase activity in vivo, as well as CTLH and CRA domains; plays role in anti-apoptosis; required for survival upon exposure to K1 killer toxin Sm-like protein ATPase stabilizing factor 0,424 Mitochondrial intermembrane space protein of unknown function; imported via the MIA import machinery; contains an unusual twin cysteine motif (CX13C CX14C 7.3 kDa subunit 9 of the ubiquinol cytochrome c oxidoreductase complex Protein required for accurate chromosome segregation during meiosis; involved in meiotic telomere clustering (bouquet formation) and telomere-led rapid prophase movements (1, 2 and see Summary Paragraph) Component of the BUB2-dependent spindle checkpoint pathway, interacts with Bfa1p and functions upstream of Bub2p and Bfa1p 0,420 YKL156W YIL097W YJR022W YDL130WA YBL107C YGR183C YPL200W YNL164C YGR046W YDR097C YBR010W YGR039W Homolog of the human GTBP protein, forms a complex with Msh2p to repair both single-base and insertion-deletion mispairs, redundant with Msh3p in repair of insertion-deletion mispairs 0,422 0,422 0,422 0,421 0,419 0,418 0,416 0,416 0,415 Histone H3 (HHT1 and HHT2 code for identical proteins) Mitochondrial protein required for cardiolipin biosynthesis; viability of null mutant is strain-dependent; mRNA is targeted to the bud; mutant displays defect in mitochondrial protein import, likely due to altered membrane lipid composition Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 Putative protein of unknown function; similar to the mouse IMPACT gene; YDL177C is not an essential gene 0,415 0,414 YLR200W Polypeptide 6 of a Yeast Non-native Actin Binding Complex, homolog of a component of the bovine NABC complex 0,413 YOL149W YBR148W YKR035WA YDR325W Decapping protein involved in mRNA degradation Spore-specific protein Rad52p inhibitor 0,411 0,411 0,410 Yeast Condensin G 0,410 YNL224C YDL177C 0,414 0,413 YDL170W YDL225W YGR187C YMR130W YGR103W YOR370C YLR144C YHL007C YPL070W YLR164W YER030W YGR068C YBR154C YOR210W YOL124C YDR235W YMR263W YDR120C YKL035W YDL178W YMR309C YDR098C-R YNL153C YLR435W YNL217W YKL092C zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type Septin homolog (putative) Hmg1\/2 protein Putative protein of unknown function; YMR130W is not an essential gene Component of several different pre-ribosomal particles; forms a complex with Ytm1p and Erb1p that is required for maturation of the large ribosomal subunit; required for exit from G0 and the initiation of cell proliferation (1, 3, 4, 5) Rab geranylgeranyltransferase regulatory subunit involved in cortical actin cytoskeleton assembly serine\/threonine protein kinase Cytoplasmic protein of unknown function containing a Vps9 domain; computational analysis of large-scale protein-protein interaction data suggests a possible role in transcriptional regulation Mitochondrial inner membrane protein of unknown function; similar to Tim18p and Sdh4p; a fraction copurifies with Sdh3p, but Shh4p is neither a stoichiometric subunit of succinate dehydrogenase nor of the TIM22 translocase; expression induced by nitrogen limitation in a GLN3, GAT1dependent manner Histone chaperone for Htz1p/H2A-H2B dimer; required for the stabilization of the Chz1p-Htz1-H2B complex; has overlapping function with Nap1p; null mutant displays weak sensitivity to MMS and benomyl; contains a highly conserved CHZ motif; protein abundance increases in response to DNA replication stress Protein proposed to regulate the endocytosis of plasma membrane proteins by recruiting the ubiquitin ligase Rsp5p to its target in the plasma membrane 25-kDa RNA polymerase subunit (common to polymerases I, II and III) RNA polymerase II subunit Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain U1 snRNP protein that shares 50\% sequence similarity with Prp39p U1 snRNP protein and has multiple copies of the crn-like TPR motif 0,409 Subunit of a histone deacetylase complex, along with Rpd3p and Sin3p, that is involved in silencing at telomeres, rDNA, and silent mating-type loci; involved in telomere maintenance N2,N2-dimethylguanosine-specific tRNA methyltransferase Uridinephosphoglucose pyrophosphorylase D-Lactate Dehydrogenase (Cytochrome) ~100 kDa cytoplasmic protein Retrotransposon TYA Gag and TYB Pol genes bovine prefoldin subunit 4 homolog (putative) Protein with a potential role in pre-rRNA processing Putative protein of unknown function; weak sequence similarity to bis (5'nucleotidyl)-tetraphosphatases; (GFP)-fusion protein localizes to the vacuole; null mutant is highly sensitive to azaserine and resistant to sodium-O-vandate GTPase-activating protein (GAP) for Rsr1p\/Bud1p 0,399 0,409 0,408 0,407 0,406 0,405 0,405 0,404 0,403 0,403 0,401 0,401 0,401 0,400 0,399 0,399 0,398 0,397 0,397 0,396 0,395 0,395 0,394 0,393 0,393 YMR256C YCL054W YDR324C Q0110 YML091C YFL062W YDR316WA YBL034C YIL139C YPR132W YOR266W YJL156C YML062C YMR081C YMR319C YMR052W YER049W YPL210C YDR194C YOL077WA Q0182 YER002W YKR026C YDR068W YOR274W YEL003W YDL208W YMR212C YHR060W YOR147W YMR070W subunit VII of cytochrome c oxidase Putative methyltransferase Subunit of U3-containing 90S preribosome and Small Subunit (SSU) processome complexes involved in production of 18S rRNA and assembly of small ribosomal subunit; member of t-Utp subcomplex involved with transcription of 35S rRNA transcript mRNA maturase bI2 subunit of mitochondrial RNase P similar to subtelomerically-encoded proteins TyA gag protein. Gag processing produces capsid proteins. 0,392 0,392 0,392 component of the 17tic spindle subunit of DNA polymerase-zeta (Pol-zeta), an enzyme whose sole function appears to be translesion synthesis 0,388 0,388 Ribosomal protein S23B (S28B) (rp37) (YS14) involved in pentamidine resistance protein Serine protease of SPS plasma membrane amino acid sensor system (Ssy1pPtr3p-Ssy5p); contains an inhibitory domain that dissociates in response to extracellular amino acids, freeing a catalytic domain to activate transcription factor Stp1p mitochondrial targeting protein involved in mitochondrial RNA splicing Low-affinity Fe(II) transport protein involved in the cell cycle Poly(rA)-binding protein involved in translation termination efficiency, mRNA poly(A) tail length and mRNA stability; interacts with Sup45p (eRF1), Sup35p (eRF3) and Pab1p; similar to prolyl 4-hydroxylases; binds Fe(III) and 2oxoglutarate component of signal recognition particle mitochondrial DEAD box RNA helicase Subunit k homolog of ATP synthase 0,386 0,385 0,384 Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data Constituent of 66S pre-ribosomal particles, involved in 60S ribosomal subunit biogenesis 34 KD alpha subunit of eIF2B Protein of unknown function, green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm transfer RNA isopentenyl transferase bovine prefoldin subunit 2 homolog (putative) HMG-like nuclear protein PHO _E_ighty _F_ive _R_equiring vacuolar H+-ATPase assembly protein Mitochondrial inner membrane protein with similarity to Mdm31p, required for normal mitochondrial morphology and inheritance; interacts genetically with MMM1, MDM10, MDM12, and MDM34 2 Cys2-His2 zinc fingers at c-terminus, glutamine and asparagine rich 0,377 0,390 0,390 0,389 0,388 0,384 0,383 0,380 0,380 0,380 0,380 0,378 0,378 0,377 0,376 0,376 0,375 0,375 0,373 0,372 0,372 0,370 0,370 YOR304W YML076C YOL061W YDL132W YDL200C YDR473C YBL026W YMR040W YDR248C YLL034C YOR195W YGR119C YLR039C YPR043W YDR002W YMR300C YOR145C YOL109W YJL096W YDR101C YEL061C YNL127W YNL088W YLR107W YPL225W YGL082W ATPase component of a two subunit chromatin remodeling complex Homodimeric Zn2Cys6 zinc finger transcription factor; binds to a weak acid response element to induce transcription of PDR12 and FUN34, encoding an acid transporter and a putative ammonia transporter, respectively Phosphoribosylpyrophosphate synthetase (ribose-phosphate pyrophosphokinase) 0,369 0,368 involved in G1 cyclin degradation 6-O-methylguanine-DNA methylase snRNP from U4\/U6 and U5 snRNPs snRNA-associated protein of the Sm class Protein of unknown function that may interact with ribosomes, based on copurification experiments; homolog of human BAP31 protein Putative gluconokinase; sequence similarity to bacterial and human gluconokinase; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; upregulated by deletion of the RNAP-II associated factor, PAF1 0,368 0,367 0,365 0,365 0,365 Putative ATPase of the AAA family, required for export of pre-ribosomal large subunits from the nucleus; distributed between the nucleolus, nucleoplasm, and nuclear periphery depending on growth conditions possible leucine zipper Contains GLFG repeats in N-terminal half and heptad repeats in C-terminal half involved in transcription of ribosomal protein genes and ribosomal RNA Ribosomal protein L43A nuclear GTPase-activating protein for Ran phosphoribosylpyrophosphate amidotransferase Essential nucleolar protein required for pre-18S rRNA processing, interacts with Dim1p, an 18S rRNA dimethyltransferase, and also with Nob1p, which is involved in proteasome biogenesis; contains a KH domain Overexpression causes resistance to Zeocin mitochondrial ribosomal protein of the large subunit Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit biogenesis; interacts directly with Alb1; responsible for Tif6 recycling defects in absence of Rei1; associated with the ribosomal export complex kinesin-related protein involved in establishment and maintenance of 17tic spindle Protein involved in recovery from cell cycle arrest in response to pheromone, in a Far1p-independent pathway; interacts with Far3p, Far7p, Far8p, Far9p, and Far10p; has similarity to the N- and C-termini of N. crassa HAM-2 topoisomerase II, Top2p localizes to axial cores in meiosis RNA EXonuclease\; part of family of 3'-to5' exonucleases. See Moser et al. 1997 Nucleic acids Res. 25:5110-5118 0,364 Protein of unknown function; may interact with ribosomes, based on copurification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; protein abundance increases in response to DNA replication stress Putative protein of unknown function; predicted prenylation/proteolysis target of Afc1p and Rce1p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YGL082W is not an essential gene 0,351 0,368 0,365 0,364 0,363 0,362 0,361 0,361 0,359 0,359 0,359 0,357 0,354 0,354 0,353 0,352 0,352 0,351 YDR060W Constituent of 66S pre-ribosomal particles, required for large (60S) ribosomal subunit biogenesis; involved in nuclear export of pre-ribosomes; required for maintenance of dsRNA virus; homolog of human CAATT-binding protein 0,350 YBR162WA YBR039W YMR284W YBR022W involved in the secretory pathway 0,347 gamma subunit of mitochondrial ATP synthase DNA binding protein Phosphatase that is highly specific for ADP-ribose 1''-phosphate, a tRNA splicing metabolite; may have a role in regulation of tRNA splicing Ubiquinol cytochrome-c reductase subunit 8 (11 kDa protein) Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR283W is not an essential gene Protein of unknown function; similar to Pbi2p; double null mutant lacking Pbi2p and Yhr138cp exhibits highly fragmented vacuoles; protein abundance increases in response to DNA replication stress Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutation is viable transcription factor, member of Ada\/Gcn5 protein complex Sm-like protein transcription factor involved in karyogamy Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; null mutant displays increased levels of spontaneous Rad52p foci; IRC23 has a paralog, BSC2, that arose from the whole genome duplication Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; null mutant displays increased levels of spontaneous Rad52p foci; IRC23 has a paralog, BSC2, that arose from the whole genome duplication Mitochondrial intermembrane space protein; mutation affects mitochondrial distribution and morphology; contains twin cysteine-x9-cysteine motifs; protein abundance increases in response to DNA replication stress 0,346 0,344 0,344 YJL166W YLR283W YHR138C YOR050C YPL254W YNL147W YCL055W YOR044W YOR055W YKL053C-A YNL175C YDL157C 0,342 0,342 0,339 0,339 0,338 0,335 0,333 0,333 0,333 0,333 nucleolar protein Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies YGR008C ATPase stabilizing factor YKR071C Description YBR063C Probable phosphopanthethein-binding protein YDR322C-A subunit e of mitochondrial F1F0-ATPase YMR311C Regulates activity of protein phosphatase 1, Glc7p, which is involved in proper chromosome segregation 0,333 0,332 YEL020W- mitochondrial inner membrane translocase A YMR158W- Dubious open reading frame unlikely to encode a protein, based on available A experimental and comparative sequence data; overlaps the verified gene ATG16/YMR159C YNL038W Protein involved in the synthesis of N-acetylglucosaminyl phosphatidylinositol (GlcNAc-PI), the first intermediate in the synthesis of glycosylphosphatidylinositol (GPI) anchors; homologous to the human PIG-H 0,328 0,332 0,332 0,329 0,329 0,329 0,326 0,325 protein YNR014W YNL218W YJL048C YCR003W YJR097W YOR203W YDL219W YGL054C YLR410WB YBL046W YDR087C YIL063C YKR100C YMR107W YER048WA YOR254C YDR051C YDL147W YDR532C Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible; YNR014W has a paralog, YMR206W, that arose from the whole genome duplication Protein with DNA-dependent ATPase and ssDNA annealing activities; involved in maintenance of genome; interacts functionally with DNA polymerase delta; homolog of human Werner helicase interacting protein (WHIP); forms nuclear foci upon DNA replication stress UBX (ubiquitin regulatory X) domain-containing protein that interacts with Cdc48p, transcription is repressed when cells are grown in media containing inositol and choline mitochondrial ribosomal protein MRPL32 (YmL32) Protein of unknown function, contains a CSL Zn finger and a DnaJ-domain; involved in diphthamide biosynthesis; ortholog human Dph4 Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps 5' end of essential DED1 gene required for translation initiation D-Tyr-tRNA(Tyr) deacylase, functions in protein translation, may affect nonsense suppression via alteration of the protein synthesis machinery; ubiquitous among eukaryotes 14 KDa protein found on ER-derived vesicles TyB Gag-Pol protein. Gag processing produces capsid proteins. Pol is cleaved to produce protease, reverse transcriptase and integrase activities. 0,324 Regulatory subunit of protein phosphatase PP4; presence of Psy4p in the PP4 complex (along with catalytic subunit Pph3p and Psy2p) is required for dephosphorylation of the histone variant H2AX, but not for dephosphorylation of Rad53p, during recovery from the DNA damage checkpoint; localization is cell-cycle dependent and regulated by Cdc28p phosphorylation; required for cisplatin resistance; homolog of mammalian R2 involved in rRNA processing nuclear protein, interacts with Gsp1p and Crm1p Transmembrane protein with a role in cell wall polymer composition; localizes on the inner surface of the plasma membrane at the bud and in the daughter cell Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources Cysteine desulfurase (Nfs1p) activator; essential for the formation of the persulfide intermediate at the desulfurase active site during pyridoxal phosphate-dependent desulfuration of cysteine; required for mitochondrial iron-sulfur cluster biosynthesi ER protein translocation subcomplex subunit Acid phosphatase involved in the non-vesicular transport of sterols in both directions between the endoplasmic reticulum and plasma membrane; deletion confers sensitivity to nickel Subunit of the regulatory particle of the proteasome Subunit of a kinetochore-microtubule binding complex with Spc105p that bridges centromeric heterochromatin and kinetochore MAPs and motors, and 0,314 0,324 0,323 0,321 0,321 0,321 0,319 0,315 0,314 0,313 0,311 0,310 0,310 0,308 0,307 0,305 0,304 0,303 YHR157W YBR290W YDR363WA YOR215C YMR269W YJL176C YJL150W YHR144C YBL028C YML019W YLR262C-A YHR121W YOR075W Q0275 YNL206C YDR476C Q0130 YBR244W YMR230W YCL066W YKL099C YPL059W YMR158W YDL243C YGR182C is also required for sister chromatid bi-orientation and kinetochore binding of SAC components mRNA is induced early in meiosis copper transporter unknown function, similar to S. pombe Dss1 0,303 0,303 0,302 Putative protein of unknown function; the authentic protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays reduced frequency of mitochondrial genome loss Nucleolar protein implicated in ribosome biogenesis; deletion extends chronological lifespan transcription factor Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data dCMP deaminase Protein of unknown function that may interact with ribosomes; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus; predicted to be involved in ribosome biogenesis Putative new 37kDa subunit of N-oligosaccharyltransferase complex Protein of unknown that associates with ribosomes; null mutant exhibits translation defects, altered polyribosome profiles, and resistance to the translation inhibitor anisomcyin; protein abundance increases in response to DNA replication stress Protein of unknown function that may function in RNA processing; interacts with Pbp1p and Pbp4p and associates with ribosomes; contains an RNAbinding LSM domain and an AD domain; GFP-fusion protein is induced by the DNA-damaging agent MMS; relative distribution to the nucleus increases upon DNA replication stress 0,301 endoplasmic reticulum t-SNARE, coprecipitates with Sec20p, Tip1p. and Sec22p Cytochrome-c oxidase subunit III, mitochondrially-coded Regulator of Ty1 Transposition - same phenotype as RTT101 - RTT105, disruption causes increase in Ty1 transposition. Isolated from the same screen as the other named RTT genes. 0,295 Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YDR476C is not an essential gene F0-ATP synthase subunit 9 (ATPase-associated proteolipid) Probable glutathione peroxidase (EC 1.11.1.9) Ribosomal protein S10B transcription factor involved in the regulation of alpha-specific genes Subunit of U3-containing Small Subunit (SSU) processome complex involved in production of 18S rRNA and assembly of small ribosomal subunit Protein with glutaredoxin activity Mitochondrial ribosomal protein of the small subunit Hypothetical aryl-alcohol dehydrogenase Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified 0,291 0,301 0,300 0,299 0,299 0,299 0,297 0,296 0,296 0,295 0,293 0,291 0,289 0,287 0,286 0,286 0,285 0,285 0,285 0,284 YHL023C YGL055W YMR014W YER105C YDR088C YMR003W YHR012W YJL168C YNL085W YNR034WA YLR395C YPL146C YJL195C YDR357C YLR038C YNL122C YIL096C YCL031C YDR515W YLR166C YIR005W YCR014C YGL055WR ORF TIM13/YGR181W Subunit of SEA (Seh1-associated), Npr2/3, and Iml1p complexes; Npr2/3 complex mediates downregulation of TORC1 activity upon amino acid limitation; SEA complex is a coatomer-related complex that associates dynamically with the vacuole; Iml1p complex (Iml1p-Npr2p-Npr3p) is required for non-nitrogen-starvation (NNS)-induced autophagy; required for Npr2p phosphorylation and Iml1p-Npr2p interaction; null mutant shows delayed meiotic DNA replication and double-strand break repair delta-9-fatty acid desaturase BUD site selection Nucleoporin similar to Nup157p and to mammalian Nup155p involved in mRNA splicing Protein of unknown function; GFP-fusion protein localizes to the mitochondria; null mutant is viable and displays reduced frequency of mitochondrial genome loss involved in vacuolar protein sorting transcription factor containing a SET domain retroviral protease signature protein Putative protein of unknown function; expression is regulated by Msn2p/Msn4p; YNR034W-A has a paralog, YCR075W-A, that arose from the whole genome duplication Cytochrome-c oxidase chain VIII Nucleolar protein; involved in biogenesis of the 60S subunit of the ribosome; interacts with rRNA processing factors Cbf5p and Nop2p; null mutant is viable but growth is severely impaired Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YJL194W/CDC6 Protein of unknown function; likely member of BLOC complex involved in endosomal cargo sorting; null mutant is sensitive to drug inducing secretion of vacuolar cargo; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm subunit VIb of cytochrome c oxidase Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YNL122C is not an essential gene Putative S-adenosylmethionine-dependent methyltransferase; associates with precursors of the 60S ribosomal subunit; predicted to be involved in ribosome biogenesis involved in rRNA processing RNA binding protein with La motif 100 kD component of the Exocyst complex\; required for exocytosis. The Exocyst complex contains the gene products encoded by SEC3, SEC5, SEC6, SEC8, SEC10, SEC15 and EXO70. Component of the U2 snRNP, required for the first catalytic step of splicing and for spliceosomal assembly; interacts with Rds3p and is required for Mer1p-activated splicing DNA polymerase IV 0,282 0,282 0,282 0,282 0,281 0,280 0,280 0,279 0,279 0,275 0,275 0,269 0,269 0,269 0,267 0,267 0,265 0,263 0,262 0,259 0,256 0,255 0,255 YBR211C YKL089W YMR132C regulator of microtubule stability centromere protein Protein of unknown function, contains sequence that closely resembles a J domain (typified by the E. coli DnaJ protein) 0,254 0,253 0,252 YDL005C YDR261C YLL009C YER133W YJL098W YKL079W YLR129W YIL027C Stoichiometric member of mediator complex Exo-1,3-b-glucanase cysteine-rich cytoplasmic protein protein phosphatase type I Sit4p-associated protein kinesin heavy chain homolog interacts with Dom3p Member of a transmembrane complex required for efficient folding of proteins in the ER; null mutant displays induction of the unfolded protein response, and also shows K1 killer toxin resistance DNA binding protein, homologous to mammalian RFX1-4 proteins similar to beta-transducin superfamily Protein involved in Rad1p/Rad10p-dependent removal of 3'-nonhomologous tails during double-strand break repair via single-strand annealing; green fluorescent protein (GFP)-fusion protein localizes to the nucleus adenylylsulfate kinase Subunit of the COMPASS (Set1C) complex, which methylates lysine 4 of histone H3 and is required in chromatin silencing at telomeres; contains a Dpy-30 domain that mediates interaction with Bre2p; similar to C. elegans and human DPY-30 Nucleolar protein, forms a complex with Noc4p that mediates maturation and nuclear export of 40S ribosomal subunits; also present in the small subunit processome complex, which is required for processing of pre-18S rRNA ubiquitin like protein Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF PEX14/YGL153W TyB Gag-Pol protein. Gag processing produces capsid proteins. Pol is cleaved to produce protease, reverse transcriptase and integrase activities. 0,249 0,248 0,248 0,246 0,244 0,243 0,242 0,241 coiled-coil protein involved in the spindle-assembly checkpoint SPOUT methyltransferase; catalyzes omega-monomethylation of Rps3p on Arg-146; not an essential gene; predicted to be involved in rRNA processing and ribosome biogenesis and in biopolymer catabolism hypoxanthine guanine phosphoribosyltransferase ubiquitin-like protein SCEI -DNA endonuclease in mitochondrial 21S rRNA intron Component of H\/ACA-box snoRNPs 0,227 0,227 Constituent of 66S pre-ribosomal particles, involved in 60S ribosomal subunit biogenesis; localizes to both nucleolus and cytoplasm subunit II of cytochrome c oxidase involved in mRNA decay TFIIS-like small Pol III subunit C11 argininosuccinate lyase 0,221 YLR176C YLR196W YAL027W YKL001C YDR469W YDL148C YIL008W YGL152C YNL054WB YGL086W YOR021C YDR399W YDR139C Q0160 YHR072WA YNL110C Q0250 YKL009W YDR045C YHR018C 0,241 0,238 0,237 0,233 0,232 0,232 0,232 0,229 0,229 0,227 0,226 0,223 0,223 0,220 0,218 0,217 0,216 YKL051W YML009C YDR468C YML026C YBL040C YNL104C YGR023W YER159C YGR090W YJL138C YLR051C YMR188C YDL140C YDL166C YMR124W YJR152W YDL058W Q0075 YER112W YHR037W YDR168W YGR168C YLR456W YPL265W YBR067C YBR048W Q0297 YDR175C YLR190W Plasma membrane protein that may act together with or upstream of Stt4p to generate normal levels of the essential phospholipid PI4P, at least partially mediates proper localization of Stt4p to the plasma membrane mitochondrial ribosomal protein MRPL39 (YmL39) tSNARE that affects a Late Golgi compartment Ribosomal protein S18B encodes the HDEL receptor required for retention of ER proteins alpha-isopropylmalate synthase (2-Isopropylmalate Synthase) acts in concert with Mid2p to transduce cell wall stress signals Transcriptional regulator which functions in modulating the activity of the general transcription machinery in vivo 0,216 Component of the small-subunit processome; required for nuclear export of tRNAs; may facilitate binding of Utp8p to aminoacylated tRNAs and their delivery to Los1p for export; conserved from yeast to mammals translation initiation factor eIF4A Essential nucleolar protein involved in the early steps of 35S rRNA processing; interacts with Faf1p; member of a transcriptionally co-regulated set of genes called the RRB regulon Mitochondrial ribosomal protein of the small subunit RNA polymerase II large subunit Essential NTPase required for small ribosome subunit synthesis, mediates processing of the 20S pre-rRNA at site D in the cytoplasm but associates only transiently with 43S preribosomes via Rps14p, may be the endonuclease for site D Protein of unknown function; GFP-fusion protein localizes to the cell periphery, cytoplasm, bud, and bud neck; interacts with Crm1p in two-hybrid assay; YMR124W is not an essential gene; predicted to have a role in organelle organization allantoate permease Integrin analogue gene intron of mitochondrial COX1, aI5-beta U6 snRNA associated protein delta-1-pyrroline-5-carboxylate dehydrogenase (putative) chaperone, involved in spindle pole body duplication and passage through START 0,199 Putative protein of unknown function; YGR168C is not an essential gene Putative pyridoxal 5'-phosphate synthase; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2; YLR456W has a paralog, YPR172W, that arose from the whole genome duplication dicarboxylic amino acid permease cell wall mannoprotein Ribosomal protein S11B (S18B) (rp41B) (YS12) Dubious open reading frame of the mitochondrial genome; unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene RPM1 protein of the small subunit of the mitochondrial ribosome Phosphorylated protein of the mitochondrial outer membrane, localizes only to mitochondria of the bud; interacts with Myo2p to mediate mitochondrial 0,168 0,168 0,213 0,212 0,211 0,208 0,206 0,206 0,199 0,197 0,194 0,193 0,189 0,188 0,185 0,181 0,173 0,173 0,170 0,169 0,168 0,165 0,163 0,161 0,151 0,149 0,140 Q0055 YDL134C Q0070 YBR054W YKL058W YOR095C YNL067W Q0050 YJL189W YKR065C Q0080 YGR084C YNL188W YHR196W YOR089C YHR029C YMR238W YMR143W YOR293W YCL025C YKL056C YAL007C YHR023W Q0065 YDR450W Q0045 YIL076W YHR071W distribution to buds; mRNA is targeted to the bud via the transport system involving She2p maturase aI2\; encodes intron-specific reverse transcriptase activity\; putative endonuclease 0,138 serine-threonine protein phosphatase 2A DNA endonuclease involved in intron homing Homolog to HSP30 heat shock protein YRO1 (S. cerevisiae) 7 Transcription factor IIA, small chain Ribose-5-phosphate ketol-isomerase Ribosomal protein L9B (L8B) (rp24) (YL11) maturase aI1\; intron-specific reverse transcriptase activity Ribosomal protein L39 (L46) (YL40) Constituent of the TIM23 complex; proposed alternatively to be a component of the import motor (PAM complex) or to interact with and modulate the core TIM23 (Translocase of the Inner mitochondrial Membrane) complex; protein abundance increases in response to DNA replication stress mitochondrially-encoded ATP synthase subunit 8 35 kDa mitochondrial ribosomal small subunit protein Localizes to the spindle pole body Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA small GTP-binding protein\; geranylgeranylated\; geranylgeranylation required for membrane association\; also involved in endocytosis post vesicle internalization 0,133 0,131 0,126 0,126 0,125 0,122 0,122 0,119 0,117 Thymidylate synthase (putative\; weak) involved in pseudohyphal growth Ribosomal protein S16A (rp61R) Ribosomal protein S10A Amino acid permease Protein that associates with ribosomes; homolog of translationally controlled tumor protein; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and relocates to the mitochondrial outer surface upon oxidative stress p24 protein involved in membrane trafficking Class II Myosin I-SceII endonuclease activity is encoded by the aI4 intron Ribosomal protein S18A cytochrome-c oxidase subunit I epsilon-COP coatomer subunit Sec28p G1\/S cyclin (weak) 0,099 0,095 0,090 0,083 0,082 0,079 0,116 0,114 0,107 0,103 0,099 0,074 0,066 0,054 0,047 0,044 0,044 0,004