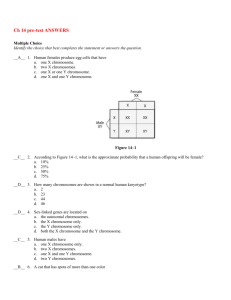
Literature_review_work
advertisement

KARYOMORPHOLOGICAL STUDIES IN TREE LEGUMES OGUNTUASE OLUWASEUN A. (B.SC PLANT SCIENCE) MATRIC. NUMBER 146574 A literature review in the department of BOTANY AND MICROBIOLOGY, Faculty of Sciences. Submitted to the School of Postgraduate Studies, University of Ibadan, in partial fulfilment of the requirement for the award of the degree of MASTER OF SCIENCE UNIVERSITY OF IBADAN. AUGUST, 2010. 1 CERTIFICATION This is to certify that the literature review of this thesis and the subsequent preparation of this thesis by Oguntuase Oluwaseun Anthony, Matriculation number 146574 were carried out under my supervision and that to the best of my knowledge this work has not been presented elsewhere for the award of any degree. ..................................................................... Dr. A. I. Adesoye (B. Sc, M. Sc, PhD. (Ibadan)) (Supervisor) 2 DEDICATION I dedicate this research work to God the Father, the Almighty; Jesus Christ, the only Son of God who is in being with the Father; and to the Holy Spirit, the Lord, the Giver of life, who with the Father and the Son is adored and glorified. 3 ACKNOWLEDGEMENT I wish to thank God for the possibility and attainment of this Master’s programme and this literature review. May His kingdom come. 4 TABLE OF CONTENT TITLE PAGE..............................................................................................................................i CERTIFICATION ....................................................................................................................ii DEDICATION............................................................................................................................iii ACKNOWLEDGEMENT..........................................................................................................iv TABLE OF CONTENTS...........................................................................................................v LIST OF TABLES......................................................................................................................ix LIST OF FIGURES....................................................................................................................x ABSTRACT................................................................................................................................xi CHAPTER ONE 1.0 INTRODUCTION...........................................................................................................1 1.1 Legume.........................................................................................................................1 1.2 Classification of Legumes.............................................................................................1 1.3 Tree Legumes................................................................................................................2 1.4 Importance of Tree Legumes.........................................................................................3 1.4.1 Degradation of Natural Systems.....................................................................4 1.4.2 Soil Reclamation and Erosion Control...........................................................4 1.4.3 Tree Legumes as Forage for Animals.............................................................5 1.4.4 Tree Legumes for Fuelwood...........................................................................6 1.4.5 Tree Legumes As Planted Forage In Cropping and Grazing Systems ...........8 5 1.4.6 Tree Legumes as Nutrition Supplements........................................................9 1.4.6 Tree Legumes as Nutrition Supplements........................................................9 1.11 Objectives of the Review.............................................................................................9 CHAPTER TWO 2.1 Introduction to Karyomorphology..........................................................................................11 2.2 Karyotypes and Morphology...................................................................................................11 2.3 Karyotypes and Karyomorphological Studies ........................................................................11 2.3.1 Plants in General.......................................................................................................11 2.3.2 In Legumes................................................................................................................12 2.4 Techniques Used in Karyomorphological Studies...................................................................13 2.4.1 The Conventional Method........................................................................................14 Plant Material.........................................................................................................14 Pretreatment..........................................................................................................14 Fixation..................................................................................................................15 Hydrolysis..............................................................................................................15 Staining..................................................................................................................15 2.5 Some New and Modern Methods...........................................................................................16 2.5.1 Flow Cytometry Method..........................................................................................16 6 2.6 Depiction of Karyotypes..........................................................................................................17 2.6.1 Classic Karyotype Cytogenetics...............................................................................18 2.6.2 Spectral Karyotype (SKY Technique)......................................................................18 2.6.3 Digital Karyotyping..................................................................................................19 2.7 Methods Used In the Analysis of Karyotypes Studies...........................................................19 2.8 Parameters Observed In Karyomorphological Studies............................................................20 CHAPTER THREE 3.0 Applications of Karyomorphological Studies..........................................................................22 3.1 Determination of Ploidy Level.....................................................................................22 3.1.1 Ploidy............................................................................................................22 3.1.2 Haplo-diploidy..............................................................................................22 3.1.3 Endopolyploidy.............................................................................................22 3.1.5 Chromosomal Polymorphism.......................................................................23 3.2 Other Uses of Karyomorphological Studies................................................................24 CHAPTER FOUR 4.0 Review of Karyomorphological Studies In Some Leguminous Trees.......................26 7 CHAPTER FIVE 5.0 Conclusion.........................................................................................................................34 5.1 Chromosome Studies In Tree Legume........................................................................34 5.2 Ploidy Level In Tree Legume......................................................................................34 REFERENCES.............................................................................................................................36 8 LIST OF TABLES 1. Chromosome Number and Ploidy Level of Some Major Tree Legumes.......................26 9 ABSTRACT Worldwide, chromosomes have been counted for many tree legumes. As very few are known for some genus there have been some extensive knowledge in some others. 169 tree legumes were reviewed among legume trees which include Abrus (1), Albizzia (2), Alhagi (1), Astragalus (1), Bauhinia (3), Cajanus (11), Dalbergia (10), Erythrina (1), Gleditsia (2), Laburnum (2), Mimosa (2), Prosopis (11), Robinia (1), Senna (1), Sesbania (1), Tamarindus (1), Acacia (60), Cassia (11), Leucaena (11), Lonchocarpus (46). The chromosome numbers and the ploidy level were majorly reviewed among other information documented at KEW Database on these genera. It was observed that most sampled tree legume have their chromosome number n = 13. The ploidy level ranges from 2 to 8. Constant chromosome number of 22 was noted in the entire Lonchocarpus trees assessed. It was also observed also that genera Prosopis and Cassia have the same chromosome number of 28. Likewise Abrus, Cajanus and Lonchocarpus have the same haploid number of chromosomes as n = 22. The genera Astragalus, Robinia, Senna, Sesbania, Erythrina, Gleditsia, Laburnum, Abrus, Albizzia, Alhagi, and Bauhinia are under-represented to justify the constancy or differences of the chromosome numbers and ploidy levels. Between the genera Cassia and Astragalus with the highest and lowest chromosome numbers respectively, there are intermediate genera to bridge the gap of relatedness among the tree legume. The types of chromosome in relation to the position of the centromere in the tree legumes have no major documentation like their chromosome numbers and ploidy levels. Other data that were not commonly documented for these tree legumes include the relative sizes of their chromosomes and the number and positions of their satellite (if any). Also not listed is the degree and distribution of heterochromatic regions of these chromosomes. 10 CHAPTER ONE 1.0 INTRODUCTION 1.1 LEGUME A legume in botanical writing is a plant in the family Fabaceae (or Leguminosae), or a fruit of these specific plants. A legume fruit is a simple dry fruit that develops from a simple carpel and usually dehisces (opens along a seam) on two sides. A common name for this type of fruit is a pod, although "pod" is also applied to a few other fruit types, such as vanilla and radish (http://en.wikipedia.org/wiki/Legume). The Fabaceae, or legumes, constitute the third largest family of flowering plants, comprising more than 650 to 700 genera and 18,000 to 20,000 species (Polhill and Raven, 1981; Doyle and Luckow, 2003; Lewis et al., 2005). The legumes are, in fact, an old family. The history of legumes is tied in closely with that of human civilization, appearing early in Asia, the Americas (the common Phaseolus bean in several varieties), and Europe (broad beans) by 6,000 BC, where they became a staple, essential for supplementing protein where there was not enough meat. For reference, the common ancestor of soybean and pea, estimated at approximately 54 million years (Lavin et al., 2005), predates by 10 million years the earliest complete primate fossil, Darwinius masillae (Franzen et al., 2009). That early small primate lived in a paratropical rain forest already richly populated by leguminous trees (Engelhardt, 1922). Legumes range from tiny herbs to giant trees, dominating many tropical rainforests. Economically, legumes represent the second most important family of crop plants after Poaceae (grass family), accounting for approximately 27% of the world's crop production. Legumes cover 180,000,000 ha and 12-15% of the arable land in the world (Graham and Vance, 2003). 11 1.2 CLASSIFICATION OF LEGUMES The legumes have traditionally been placed into the three subfamilies: the Papilionoideae, with approximately 70% of species; the Mimosoideae, with approximately 15%; and the remainder in the Caesalpinoideae—though this last subfamily is now known to be comprised of a collection of early diverging legume taxa. Recent results indicate that the Caesalpinoideae is actually paraphyletic. (Doyle and Luckow, 2003; Lewis et al., 2005). According to Young et al., 2003, most cultivated legumes are found within the Papilionoideae, the subfamily with largest total number of genera. This subfamily contains nearly all economically important crop legumes, including soybean (Glycine max), peanut (Arachis hypogaea), mungbean (Vigna radiata), chickpea (Cicer arietinum), lentil (Lens culinaris), common bean (Phaseolus vulgaris), pea (Pisum sativum), and alfalfa (Medicago sativa). With the notable exception of peanut, all these important crop legumes of agronomic interest fall into two Papilionoid clades, namely, Galegoid (including the genera Melilotus, Trifolium, Medicago, Pisum, Vicia, Lotus, Cicer and Lens) and Phaseoloid (including the genera Phaseolus, Vigna, Glycine and Cajanus), which are often referred to as cool season and tropical season legumes, respectively. Doyle and Luckow, 2003, and Lewis et al., 2005 also reported that the four large subdivisions in the Papilionoideae are the galegoid, millettioid, dalbergioid, and genistoid clades. Despite their close phylogenetic relationships, crop legumes differ in their base chromosome number and ploidy level (Table I). Nevertheless, earlier studies indicated that members of the Papilionoideae subfamily exhibited extensive genome conservation based on comparative genetic mapping (Weeden et al., 1992; Menancio-Hautea et al., 1993). The papilionoid subfamily includes most crop legumes and the major model legume species, and thus is the taxonomic space across which much of legume comparative genomics and translational genomics will take place (Cannon et al., 2009). The 12 papilionoid origin is dated at approximately 59 million years (Lavin et al., 2005). The galegoid clade contains the robinioid clade, with birdsfoot trefoil and several allied forage and tree legumes (including Sesbania, and Robinia, e.g. the black locust tree). Of course, other legume genera are important to people — forest and lumber trees in Mimosoideae, such as Acacia and Albizia; other forage crops and shade trees in the Papilionoideae, such as Robinia (locust bean); and finally, important ornamental, timber and even food plants in Caesalpinoideae, such as Poinciana (flame tree) and Tamarindus (the source of tamarind fruit). 1.3 TREE LEGUMES Until recently, according to Gutteridge and Shelton (1994), tree legumes were largely neglected by researchers because their utilisation and management fell between the disciplines of forestry and pasture agronomy. They are now receiving increased research attention because of their multipurpose value and some distinctive features which set them apart from herbaceous legumes. Tree legumes can be regarded as truly multipurpose trees for agriculture in respect of their special characteristics which include 1. They are usually long-lived and low maintenance, and therefore enhance the sustainability of farming systems, 2. They provide high quality forage for feeding of livestock, 3. They stabilise sloping lands against erosion because of their deep-rooted habit, 4. They supply nitrogen-rich mulch for cropping systems, 5. They can be used to colonise and rehabilitate adverse environments, e.g. saline or arid locations, 13 6. They provide a source of timber and firewood for either domestic or industrial use, 7. They are used in farming systems as living fences, as shade trees for plantation crops, and as living trellises for climbing crops, and 8. They are a source of fruit and vegetables for human consumption. 1.4 IMPORTANCE OF TREE LEGUMES 1.4.1 Degradation of Natural Systems Unfortunately, due in large part to over-exploitation by people and livestock, valuable tree and shrub resources over vast areas in arid and semiarid regions have been destroyed in the last few decades. In these areas, it is important that management practices are adopted which foster the wise use of diminishing tree and shrub resources. Livestock access should be restricted, and pruning and harvesting of products performed on a rotational basis, to ensure time for regeneration. In some cases, the presence of tree legumes has contributed to the degradation of the landscape. Animals can be maintained long after the loss of palatable perennial grass species due to drought or overgrazing, by feeding the foliage of hardy tree species. Vast areas of South-western Queensland and East Africa have been degraded in this way. 1.4.2 Soil Reclamation and Erosion Control The restoration and maintenance of soil fertility is a basic and critical environmental problem. It is especially serious in tropical and subtropical regions where many soils lack plant nutrients and organic matter and intense rainfall erodes vulnerable top soil. The nitrogen fixing ability of tree legumes allows them to grow on difficult sites subject to erosion, low fertility or 14 other adverse soil conditions. Once established, they can create conditions favourable for the growth of other species leading to a balanced plant ecosystem. Tree legumes are a good source of organic matter for green manure. Their dry foliage contains 2.5-5.5% nitrogen and leaf material incorporated into the soil improves fertility, moisture and nutrient retention and general filth. At the same time, by improving soil structure, erosion can be retarded. The extensive root systems of tree legumes enable them to adapt to steeply sloping sites unsuited to conventional cropping or grazing thus stabilising the sites from erosion and providing a measure of production which would not otherwise exist. The Sloping Agricultural Land Technology (SALT) developed in the Philippines (Tacio et al., 1987) is a prime example of the use of tree legumes in substantially reducing soil erosion and restoring moderately degraded hilly lands to a profitable farming system. 1.4.3 Tree Legumes as Forage for Animals Browse has been defined as the leaves, shoots and sprouts including tender twigs and stems of woody plants which are cropped to a varying extent by domestic and wild animals. It should be extended to include the fruit, pods and seeds which provide valuable feed, especially if the tree is deciduous. Trees and shrubs have provided valuable forage to man's herbivorous animals probably since the time of their domestication (Robinson 1985). At least 75% of the shrubs and trees of Africa serve as browse plants and many of these are leguminous (Skerman 1977). McKell (1980) pointed out that shrubs and trees are the most visible plant forms in many landscapes, yet have been neglected in most scientific research. Much research effort has concentrated on methods for their eradication. In some arid and semiarid climates, livestock would not exist without browse species to supply feed. Many tree legume species have evolved in semiarid regions alongside herbivorous animals and therefore have developed means of 15 protection against browsing or grazing. Among the protective devices are thorns, toxins, fibrous foliage and height of tree crowns (Brewbaker 1986). Thorns characterise many woody legumes and are particularly prevalent on juvenile plants. Toxins are of two general types, those which deter feeding and those which poison the animal. The use of naturally occurring browse species is a vital component of livestock production systems in many regions of the world. In the Sahelian savannahs in Africa from Senegal to the Sudan, Faidherbia albida is a native leguminous species which is extremely important both in providing forage for livestock and in enhancing soil fertility for crops. Prosopis species provide forage for the sheep and cattle industries of the arid subtropical plains of Brazil, Argentina, Uruguay and Northern Chile. Prosopis chilensis contributes regular cattle feed in northwest Argentina and central Chile while P. tamarugo, a native of Chile's northern plateau, is the only tree that survives on the arid salt flats producing the only available forage, timber and fuelwood in that region. In South-western Queensland and northern New South Wales, mulga (Acacia aneura) occurs naturally often in monospecific stands and is used as a drought reserve for grazing sheep. Under natural conditions, a large proportion of the foliage of tree species will be out of reach of grazing animals so utilisation can be manipulated by cutting or lopping to make it available when needed. Sometimes natural leaf fall through senescence is an important day-to-day component of the diet of some grazing animals. In Africa, goats thrive on the leaf fall of Acacia melliflora (Dougall and Bogden 1958). 1.4.4 Tree Legumes for Fuelwood In many of these intensive cropping areas, tree legumes are planted not only for their forage but also for firewood, green manure and other uses. It has been estimated (Eckholm 1975) 16 that at least half the timber cut in the world is used as a fuel for cooking and heating. Approximately 2 billion people derive at least 90% of their energy requirements from wood and charcoal while a further 1.5 billion meet at least 50% of their requirements this way. This essential resource, however, is seriously threatened. If the pace of tree planting around the world is not greatly accelerated, at least 500 million people will be without fuelwood for their minimum cooking and heating needs by the end of the century. Tree legumes offer a partial solution to the fuelwood crisis. Of the 88 species recommended for fuelwood production by the National Academy of Sciences (1980, 1983), almost half were tree legumes. Tree legumes also meet many of the characteristics which are considered desirable in fuelwood species which include: 1. Rapid growth, 2. Nitrogen fixing ability, 3. Ease of establishment, 4. Ability to coppice, 5. Wood of high calorific value, 6. Wood which burns without sparks or toxic smoke, 7. Ability to grow well in a wide range of environments including difficult sites, 8. Multipurpose nature. Tree legumes have been used successfully in sustained fuelwood production systems. In the early 1920s, in the Paliparan area of the Philippines, Leucaena leucocephala was planted over a large area of unproductive Imperata cylindrica grassland. Since then it has yielded on average 20 m3 of fuelwood per hectare per year and is still the main source of fuel for the city of Laguna (NAS, 17 1980) although recent damage by the Leucaena psyllid has reduced the production of wood. Other tree legumes that are highly regarded as fuelwood species include Acacia auriculiformis, A. saligna, A. senegal, A. tortilis, Calliandra calothyrsus, Cassia siamea, Pithocellobium dulce and Prosopis spp. 1.4.5 Tree Legumes As Planted Forage In Cropping and Grazing Systems As well as naturally occurring stands, tree legumes are often planted specifically for forage both in extensive grazing systems and in association with crops. In many of the more intensive agricultural areas of Asia and Africa, where livestock are raised in small numbers by smallholder farmers, tree legumes are planted as 'forage banks' on unused land along field borders or fence lines, on rice paddy bunds or in home gardens. These areas are usually harvested under a 'cut-and-carry' system and are a principal source of high quality forage used to supplement low quality roughages such as crop residues. Productivity from these areas can be quite high. In the Batangas region of the Philippines, a 2 ha area of Leucaena leucocephala grown in association with the fruit tree Anona squamosa was able to supply the forage requirements of 20 growing cattle over a 6 month period (Moog 1985). At Ibadan in Nigeria, Reynolds and Atta-Krah (1986) suggested that the surplus foliage produced over a year from 1 ha of Leucaena leucocephala and Gliricidia sepium planted at 4 m intervals in an alley cropping system could be used as a supplement to provide half the daily forage requirements for 29 goats. In the more extensive grazing areas of Australia, southern Africa and South America, tree legumes are increasingly being planted in association with improved grasses to increase carrying capacity and productivity of grazing cattle. In central Queensland, over 20,000 ha have been sown to Leucaena leucocephala in the past 10 years. The Leucaena is sown in wide spaced rows 18 4-10 m apart and an improved grass such as green panic (Panicum maximum var. trichoglume), Rhodes grass (Chloris gayana), buffer grass (Cenchrus ciliaris) or signal grass (Brachiaria decumbens) sown between the Leucaena rows. A high stocking rate (up to 3-4 animals/ha) and liveweight gain (up to 1 kg/head/day) can be achieved with this system. A record liveweight gain of 1,442 kg/ha for cattle grazing a grass/legume pasture was achieved on an irrigated leucaena/pangola grass mixture in the Ord River District of north Western Australia (Jones 1986). Other tree legume species that are being investigated for use in extensive grazing systems include Calliandra calothyrsus, Albizia chinensis, Cajanus cajan, Gliricidia sepium and Sesbania sesban. 1.4.6 Tree Legumes as Nutrition Supplements On a worldwide basis, legumes contribute about one-third of humankind's protein intake. Legumes also accumulate natural products (secondary metabolites) such as isoflavonoids that are beneficial to human health through anticancer and other health-promoting activities (Dixon and Sumner, 2003). The nutritional quality of tree legumes varies from excellent (Leucaena leucocephala) to quite poor (most Australian Acacia species). Poor quality can be due to tannins which reduce the digestibility of both herbage and protein. The presence of tannins is often evident as brownish, reddish tinges in juvenile growth. Another reason for poor quality is that some species have phyllodes (expanded and flattened leaf petioles) instead of compound pinnate or bipinnate leaves which are very high in fibre and therefore of low digestibility, e.g. the Australian acacias. Forage from tree legumes is often used as a buffer to overcome feed gaps that arise from seasonal fluctuations in the productivity of other feed sources. For example, grasses and other herbs may die when upper soil layers lose their moisture but the deep-rooted trees exploit moisture at depth and continue to grow. During the dry season or in times of drought, 19 trees provide green forage rich in protein, minerals and vitamins while the herbaceous cover provides only poor quality straw. Tree legumes have an important role in many agricultural production systems throughout the world as they can be used in a multitude of ways. The aim of this review, therefore, is to look at the genetic status of these multipurpose trees as documented by several researchers through their karyomorphological studies. This review also looked into how these karyomorphological studies are being carried out, some notable results among the legume tree, and the areas of application of karyomorphological data. 20 CHAPTER TWO 2.1 INTRODUCTION TO KARYOMORPHOLOGY This is the combine study of the karyotype and chromosome morphology of organisms. As suggested by Mhinana et al., 2010, these studies could be used as an additional tool in the classification of organism. 2.2 KARYOTYPES AND MORPHOLOGY A karyotype is the number and appearance of chromosomes in the nucleus of a eukaryote cell (White, 1973; Stebbins, 1950). The term is also used for the complete set of chromosomes in a species, or an individual organism. Karyotypes describe the number of chromosomes, and what they look like under a light microscope. Attention is paid to their length, the position of the centromeres, any differences between the sex chromosomes, and any other physical characteristics (King et al., 2006). Morphology is a branch of bioscience dealing with the study of the form and structure of organisms and their specific structural features (http://en.wikipedia.org/wiki/morphology). Therefore juxtaposing with the knowledge of karyotye, karyomorphological studies focuses on the morphological features of chromosomes. This reveals chromosomes that are metacentric, submetacentric, intercentric, chromocenter, proximal, interstitial, acrocentric, telocentric types. This is majorly determined by the position of their centromere. 2.3 KARYOTYPES AND KARYOMORPHOLOGICAL STUDIES 2.3.1 Plants in General – Chromosomes were first observed in plant cells by Karl Wilhelm von Nägeli in 1842. Their behavior in animal (salamander) cells was described by Walther 21 Flemming, the discoverer of mitosis, in 1882. The name was coined by another German anatomist, von Waldeyer in 1888. The next stage took place after the development of genetics in the early 20th century, when it was appreciated that the set of chromosomes (the karyotype) was the carrier of the genes. Levitsky (1924, 1931) seems to have been the first to define the karyotype as the phenotypic appearance of the somatic chromosomes, in contrast to their genic contents. The subsequent history of the concept can be followed in the works of Darlington (1939) and White (1973a; 1973b). The study of whole sets of chromosomes is sometimes known as karyology. The chromosomes are depicted (by rearranging a microphotograph) in a standard format known as a karyogram or idiogram: in pairs, ordered by size and position of centromere for chromosomes of the same size. The basic number of chromosomes in the somatic cells of an individual or a species is called the somatic number and is designated 2n. In normal diploid organisms, autosomal chromosomes are present in two copies. There may, or may not, be sex chromosomes. Polyploid cells have multiple copies of chromosomes and haploid cells have single copies. 2.3.2 In Legumes – The chromosome maps of some legumes such as soybean (Glycine max), Lotus (Lotus japonicus), and red clover (Trifolium pratense) have been reviewed by Ohmido et al., 2007. Papilionoideae contains the majority of pulse crops such as pea (Pisum sativum, 2n = 14, 5000 Mb), alfalfa (Medicago sativa, 2n =16, 1600 Mb), and soybean (Glycine max (L.) Merr., 2n = 40, 1100 Mb). These species, Glycine max and Lotus japonicus, have relatively small chromosomes and therefore are difficult to exploit for chromosome studies. Nevertheless, the identification of individual chromosomes became feasible, and chromosome maps have been developed applying image analysis and fluorescence in-situ hybridization. For Lotus japonicus, e.g. detailed chromosome maps have been developed using the information of genetic linkage 22 maps. For legume species with large chromosomes, such as Vicia faba (2n = 12) and Pisum sativum (2n =14), it is possible to use ordinary karyotyping and/or banding methods for chromosome identification (Ohmido et al., 2007). A comprehensive survey of the molecular and cytogical features of the chromosome complement was provided for V. faba based on FISH and various Giemsa and fluorescence banding patterns (Fuchs et al., 1998a). In the case of legumes with small chromosomes, identification of individual chromosomes and their centromeric positions is difficult, especially after condensating pretreatment with colchicine, 8-hydroxyquinoline or cold water. The Chromosome Image Analyzing System (CHIAS) for small chromosomes makes use of distinct stainability along mitotic prometaphase chromosomes, due to uneven condensation, a feature specific to small plant chromosomes (Fukui & Iijima 1991). The density profiles at the center line of both chromatids (mid-rib line) of prometaphase chromosomes allowed establishment of the first chromosome maps of several legumes with small chromosomes (Yanagisawa et al., 1991; Ito et al., 2001; Sato et al., 2005). 2.4 TECHNIQUES USED IN KARYOMORPHOLOGICAL STUDIES 2.4.1 The Conventional Method Many different methods of making squash preparations for chromosome studies have been described. These involve the application of various chemical and physical agents for the pretreatment and fixation of the dividing cells. A good technique should permit the chromosomes to become well spread during the squashing, and, at the same time, the fixation should bring out the details of chromosome morphology as distinctly as possible. Well spread chromosomes with 23 a clear morphology are essential for karyotype studies and chromosome measurements (Östergren and Heneen, 2009). For other species, various types of pretreatment may be more useful. The pretreatment to be used depends not only on the species studied but also on the special aspect of chromosome cytology in which the investigator is interested. When it is essential to get the chromosomes well separated during the squashing, it is desirable to have them strongly contracted. On the other hand, when more interest is focused on morphological landmarks, such as satellites and secondary constrictions, it is often useful to have the chromosomes more weakly contracted (Östergren and Heneen, 2009). 2.4.2 Plant materials Chromosome observation is usually carried out in cells of the root tips (Lv et al., 2009). Some preparation is made from the shoot tips of some other plants. Some of these plants are cultured in water in a greenhouse (Lv et al., 2009), Petri dishes (Mansion and Zeltner, 2004), experimental pots or in the field. 2.4.3 Pretreatment Pre-treatment of cells helps to arrest the dividing cells at metaphase by interfaring with the formation of spindle fibre. This treatment does not really kill the cell but limits the completion of the cell division. The root tips are mostly collected from young plants in the morning and pretreated with 0.002 mol/L 8-hydroxyquinoline solution from few minutes to several hours depending on the type of plant used. Other pretreatment agents used include oxyquinoline (Tjio and Levan, 1950), alpha bromonaphthalene (Schmuck and Kostoff, 1935), Colchicine (O’Mara, 1939), paradichlorobenzene (Meyers, 1945) and cold water (Hill and Myers, 1945). Besides inducing an increased contraction of the chromosomes, oxyquinoline causes an inactivation of 24 the spindle and an increase in the viscosity of the cytoplasm (Tjio and Levan, 1950; Stalfelt, 1950). 2.4.4 Fixation This treatment helps to kill the cell in such a way that all the stages of division, in which the cell is undergoing, remain as they are. The most common fixating agent is Carnoy’s solution. It is prepared using ethanol and acetic acid in ratio 3 to 1 respectively. The treatment time for fixation also vary depending on the plant used. 2.4.5 Hydrolysis This is the stage at which the plant parts used for chromosome studies are softened for easy maceration or squashing. The time and temperature for hydrolysis is mostly determined by the researcher in relation to the plants under investigation. Mostly prepared concentration for hydrolysis is 1N of Hydrochloric acid as recently used by Tabur and Oney (2009) among many other researchers. 2.4.6 Staining This is among the major stages for chromosome studies because it helps to differentiate the chromosomes from their immediate cell environment. Stains used for chromosome studies include Feulgen (Tabur and Oney, 2009), Aceto-carmine (Gill and Obembe, 1991; Medina and Conagin, 1964), Carbol Fuchsin solution (Li, 1982), Giemsa technique (Guerra, 1983), Leucobasic fushin (Badr and Gasim, 1992). 25 2.5 SOME NEW AND MODERN METHODS Chromosome squashes are time-consuming and are rapidly becoming a lost art (Goldblatt 2007). Fortunately, with recent refinements in the accuracy and cost efficiency of flow cytometry, researchers now initiate investigations into chromosome differences to detect intraspecific cytotype variation (Murray and Young, 2001; Kron et al., 2007). 2.6 DEPICTION OF KARYOTYPES According to Gustashaw (1991), Cytogenetics employs several techniques to visualize different aspects of chromosomes 1. G-banding is obtained with Giemsa stain following digestion of chromosomes with trypsin. It yields a series of lightly and darkly stained bands - the dark regions tend to be heterochromatic, late-replicating and AT rich. The light regions tend to be euchromatic, early-replicating and GC rich. This method will normally produce 300-400 bands in a normal, human genome. 2. R-banding is the reverse of G-banding (the R stands for "reverse"). The dark regions are euchromatic (guanine-cytosine rich regions) and the bright regions are heterochromatic (thymine-adenine rich regions). 3. C-banding: Giemsa binds to constitutive heterochromatin, so it stains centromeres. 4. Q-banding is a fluorescent pattern obtained using quinacrine for staining. The pattern of bands is very similar to that seen in G-banding. 5. T-banding: visualize telomeres. 26 6. Silver staining: Silver nitrate stains the nucleolar organization region-associated protein. This yields a dark region where the silver is deposited, denoting the activity of rRNA genes within the NOR. 2.6.1 Classic Karyotype Cytogenetics In the "classic" (depicted) karyotype, a dye, often Giemsa (G-banding), less frequently Quinacrine, is used to stain bands on the chromosomes. Giemsa is specific for the phosphate groups of DNA. Quinacrine binds to the adenine-thymine-rich regions. Each chromosome has a characteristic banding pattern that helps to identify them; both chromosomes in a pair will have the same banding pattern. Karyotypes are arranged with the short arm of the chromosome on top, and the long arm on the bottom. Some karyotypes call the short and long arms p and q, respectively. In addition, the differently stained regions and sub-regions are given numerical designations from proximal to distal on the chromosome arms (Shaffer and Tommerup, 2005). 2.6.2 Spectral Karyotype (SKY Technique) Spectral karyotyping is a molecular cytogenetic technique used to simultaneously visualize all the pairs of chromosomes in an organism in different colors. Fluorescently labeled probes for each chromosome are made by labeling chromosome-specific DNA with different fluorophores. Because there are a limited number of spectrally-distinct fluorophores, a combinatorial labeling method is used to generate many different colors. Spectral differences generated by combinatorial labeling are captured and analyzed by using an interferometer attached to a fluorescence microscope. Image processing software then assigns a pseudo color to each spectrally different combination, allowing the visualization of the individually colored chromosomes (Schröck et al., 1996) This technique is used to identify structural chromosome 27 aberrations in cancer cells and other disease conditions when Giemsa banding or other techniques are not accurate enough. 2.6.3 Digital Karyotyping Digital karyotyping is a technique used to quantify the DNA copy number on a genomic scale. Short sequences of DNA from specific loci all over the genome are isolated and enumerated (Wang et al., 2002). This is method is also known as virtual karyotyping. 2.7 METHODS USED IN THE ANALYSIS OF KARYOTYPES STUDIES For karyotypes studies, idiograms are constructed for each standard karyotypes. The measurements of the short and long chromosome arms, and the absolute chromosome length is usually performed on an Amplival microscope at a very high magnification (<2000) (Pavlova and Tosheva, 2004). The karyotype asymmetry can easily be described using the method of Romero Zarco (1986). The latter is expressed by two numerical parameters A1 (intrachromosomal asymmetry index) and A2 (interchromosomal asymmetry index). Karyotype asymmetry for the relations between the chromosome arms has been estimated for every sample using the following equation: A1 = 1- (Sbi / Bi) n where A1 – intrachromosomal asymmetry index, ranging from zero to one; n – number of homologous chromosome pairs; bi − average length for short arms in every homologous chromosome pair; Bi − average length for long arms in every homologous chromosome pair. 28 Karyotype asymmetry due to relations between sizes of different chromosomes has been estimated using Pearson’s dispersion coefficient A2 = S /ξ S − standard deviation where ξ − the mean of chromosome length for each sample. The interchromosomal asymmetry index A2 is independent from the chromosome size, chromosome number and has no units. This method is appropriate when there are only slight differences in the karyotype asymmetry. The variations in chromosome size and type of a given species observed in natural conditions, and the changes in the structural type of chromosomes in the evolutionary process indicate that the karyotype is most susceptible to the environmental factors resulting in genotype changes. The karyotype symmetry is a reflection of the mutation process (Pavlova and Tosheva, 2004). 2.8 PARAMETERS OBSERVED IN KARYOMORPHOLOGICAL STUDIES Some six different characteristics of karyotypes are usually observed and compared according to Stebbins (1971) 1. Differences in absolute sizes of chromosomes. Chromosomes can vary in absolute size by as much as twenty-fold between genera of the same family: Lotus tenuis and Vicia faba 29 (legumes), both have six pairs of chromosomes (n=6) yet V. faba chromosomes are many times larger. This feature probably reflects different amounts of DNA duplication. 2. Differences in the position of centromeres. This is brought about by translocations. 3. Differences in relative size of chromosomes can only be caused by segmental interchange of unequal lengths. 4. Differences in basic number of chromosomes may occur due to successive unequal translocations which finally remove all the essential genetic material from a chromosome, permitting its loss without penalty to the organism (the dislocation hypothesis). 5. Differences in number and position of satellites, which (when they occur) are small bodies attached to a chromosome by a thin thread. 6. Differences in degree and distribution of heterochromatic regions. Heterochromatin stains darker than euchromatin, indicating tighter packing, and mainly consists of genetically inactive repetitive DNA sequences. A full account of a karyotype may therefore include the number, type, shape and banding of the chromosomes, as well as other cytogenetic information. According to White (1973b), variation is often found: 1. Between the sexes 2. Between the germ-line and soma (between gametes and the rest of the body) 3. Between members of a population (chromosome polymorphism) 4. Geographical variation between races 5. Mosaics or otherwise abnormal individuals. 30 CHAPTER THREE 3.0 APPLICATIONS OF KARYOMORPHOLOGICAL STUDIES 3.1 DETERMINATION OF PLOIDY LEVEL 3.1.1 Ploidy is the number of complete sets of chromosomes in a cell. Polyploidy, where there are more than two sets of homologous chromosomes in the cells, occurs mainly in plants. It has been of major significance in plant evolution according to Stebbins (Stebbins, 1940, 1950; Comai, 2005; Adams and Wendel, 2005). The proportion of flowering plants which are polyploid was estimated by Stebbins to be 30-35%, but in grasses the average is much higher, about 70% (Stebbins, 1970). Polyploidy in lower plants (ferns, horsetails and psilotales) is also common, and some species of ferns have reached levels of polyploidy far in excess of the highest levels known in flowering plants. Polyploidy in animals is much less common, but it has been significant in some groups (Gregory and Mable, 2005). Polyploid series in related species which consist entirely of multiples of a single basic number are known as euploid. 3.1.2 Haplo-diploidy is where one sex is diploid, and the other haploid. It is a common arrangement in the Hymenoptera, and in some other groups. 3.1.3 Endopolyploidy occurs when in adult differentiated tissues the cells have ceased to divide by mitosis, but the nuclei contain more than the original somatic number of chromosomes (White, 1973c). In the endocycle (endomitosis or endoreduplication) chromosomes in a 'resting' nucleus undergo reduplication, the daughter chromosomes separating from each other inside an intact nuclear membrane according to Lilly and Duronio (2005). In many instances, endopolyploid nuclei contain tens of thousands of chromosomes (which cannot be exactly counted). The cells do not always contain exact multiples (powers of two), which is why the 31 simple definition 'an increase in the number of chromosome sets caused by replication without cell division' is not quite accurate. This process (especially studied in insects and some higher plants such as maize) may be a developmental strategy for increasing the productivity of tissues which are highly active in biosynthesis (Edgar and Orr-Weaver, 2001). The phenomenon occurs sporadically throughout the eukaryote kingdom from protozoa to man; it is diverse and complex, and serves differentiation and morphogenesis in many ways (Nagl, 1978). 3.1.4 Aneuploidy is the condition in which the chromosome number in the cells is not the typical number for the species. This would give rise to a chromosome abnormality such as an extra chromosome or one or more chromosomes lost. Abnormalities in chromosome number usually cause a defect in development. Down syndrome and Turner syndrome are examples of this. Aneuploidy may also occcur within a group of closely related species. Classic examples in plants are the genus Crepis, where the gametic (= haploid) numbers form the series x = 3, 4, 5, 6, and 7; and Crocus, where every number from x = 3 to x = 15 is represented by at least one species. Evidence of various kinds shows that that trends of evolution have gone in different directions in different groups (Stebbins and Ledley, 1972). Closer to home, the great apes have 24x2 chromosomes whereas humans have 23x2. Human chromosome 2 was formed by a merger of ancestral chromosomes, reducing the number (Ijdo et al., 1991). 3.1.5 Chromosomal Polymorphism According to Ford (1975), some animal species are polymorphic for chromosome fusions or dissociations. When this happens, the chromosome number is variable from one individual to another. Well-researched examples are the ladybird beetle Chilocorus stigma, some mantids of the genus Ameles, the European shrew Sorex araneus. There is some evidence from the case of 32 the mollusc Thais lapillus (the dog whelk) on the Brittany coast, that the two chromosome morphs are adapted to different habitats (White, 1973d). Chromosome observation has been used in many plants to identify chromosomal ploidy (Nathewet et al., 2007; Kazuo and Maltide, 1993; Nankui and Paul, 2003; Li et al., 2008; Ramon and Manuel, 2004; Brutovska et al., 2000; Agnieszka et al., 2006; Kazuo et al., 2000). Knowledge of the chromosomal ploidies of germplasms do provide theoretical basis for crossbreeding (Lv et al., 2009). Polyploid formation is associated with extensive chromosome restructuring (Ahn and Tanksley, 1993; Reinisch et al., 1994; Song et al., 1995a, 1995b; Lagercrantz and Lydiate, 1996; Brubaker et al., 1999) and possibly with retrotransposon amplification (Matzke and Matzke, 1998; Zhao et al., 1998). Identification of different polyploid populations can be used to establish seed-quarantine zones, in opposition to seed-transfer zones, as insurance that incompatible cytotypes will not be mixed (Severns and Liston, 2008). 3.2 Other Uses of Karyomorphological Studies Karyotype concept has been extensively used in characterizing and distinguishing chromosomes of various organisms (Ayşe and Haşim, 2005). Future investigation into the patterns of variation of intraspecific chromosome number will provide a safeguard for conservation of rare plants in an age of restoration, contribute important information to a relatively neglected topic in plant population and landscape genetics (Levin, 2002; Halverson et al., 2008), and enhance understanding of plant speciation (Soltis et al., 2007). A karyological study is significant in determining taxonomical position of organisms (mosses) and it seems to be effective for analyzing Antarctic mosses which are usually sterile (Kanda and Okada, 1993). Thus, the information on the karyomorphology is useful for generic 33 comparisons and might be of use to support the distinctness of species (Kazuo and Hiroshe, 2000). Karyotypes can also be used for many purposes; such as, to study chromosomal aberrations, cellular function, and to gather information about past evolutionary events. Karyomorphological studies in plant populations usually show similarities or differences in their chromosomal morphology. It also provides additional data on the variation pattern within populations for the solution of taxonomical problems and successful attempts for selection. 34 CHAPTER FOUR 4.0 RESULT OF KARYOMORPHOLOGICAL STUDIES IN SOME LEGUMINOUS TREES Worldwide, chromosomes have been counted for many tree legumes. As very few are known for some genus there have been some extensive knowledge in some others. Only 26 species of Sophora have had their chromosome numbers reported (Stiefkens et al., 2001). Several chromosome numbers reported for this genus include: n = 9, 18, 11, and 2n = 18, 28, 36, 16, 32, 22, 54, in decreasing order of frequency in each case (cf. Federov, 1974; Goldblatt, 1981a,b,c, 1984, 1988, 1990, 1991, 1994, 1996, 1998). In Table 1 below, the genera that were reviewed among legume trees include Abrus (1), Albizzia (2), Alhagi (1), Astragalus (1), Bauhinia (3), Cajanus (11), Dalbergia (10), Erythrina (1), Gleditsia (2), Laburnum (2), Mimosa (2), Prosopis (11), Robinia (1), Senna (1), Sesbania (1), Tamarindus (1), Acacia (60), Cassia (11), Leucaena (11), Lonchocarpus (46). These are 169 tree legumes documented at KEW Datadase (http://www.rbdkew.org.uk/cval/homepage.html). TABLE 1: CHROMOSOME NUMBER AND PLOIDY LEVEL OF SOME MAJOR TREE LEGUMES Genus Species Abrus precatorious Albizzia amara Albizzia lucida Alhagi Authority Chromosome no. 22 Ploidy level 2 Boiv. 26 Benth. Reference Life cycle Angiosperm group 2 Al-Turki et al., (2000) Ohri, 1996 P Eudicot 26 2 Ohri, 1996 Eudicot camelorum 16 + 2B 2 Astragalus abyssinicus 16 2 Bauhinia Bauhinia tomentosa purpurea L. L. 28 28 2 2 Bauhinia hookeri 26 Cajanus cajan (F.Muell.) Pedley (L.) Millsp. Al-Turki et al., (2000) Al-Turki et al., (2000) Ohri, 2002 P Ohri and P Kumar, 1986 Ohri, 2002 P 22 2 35 P Bennett and P Smith, 1976 Eudicot Eudicot Eudicot Eudicot Cajanus Cajanus Cajanus goensis scarabaeoides volubilis 22 22 22 2 2 2 Ohri, 1996 Ohri, 1996 Ohri, 1996 P B P Eudicot Eudicot Eudicot 22 2 Ohri, 1996 P Eudicot 22 2 Ohri, 1996 B Eudicot 22 2 Ohri, 1996 P Eudicot 22 2 Ohri, 1996 P Eudicot 22 2 Eudicot 22 2 22 2 Ohri and Singh, B 2002 Ohri and Singh, A 2002 Ohri and Singh, P 2002 lanceolaria Dalz. (L.) Thou. (Blanco) Blanco (W. & A.) Maesen (Benth.) Maesen (Haines) Maesen (F.Muell.) Maesen (Benth.) Maesen (Wight. & Am.) Maesen (Benth. ex Bak.) Maesen Lin. f. Cajanus lineatus Cajanus platycarpus Cajanus cajanifolius Cajanus acutifolius Cajanus mollis Cajanus albicans Cajanus sericeus Dalbergia 20 2 P Eudicot Dalbergia sissoo Roxb. 20 2 P Eudicot Dalbergia latifolia Roxb. 20 2 P Eudicot Dalbergia sissoides Grah. 20 2 P Eudicot Dalbergia volubilis Roxb. 20 2 P Eudicot Dalbergia horrida Dennst. 20 2 P Eudicot Dalbergia paniculata Roxb. 20 2 P Eudicot Dalbergia rubiginosa Roxb. 20 2 P Eudicot Dalbergia malabarica Prain. 20 2 P Eudicot Dalbergia melanoxylon Guill & Perr. 20 2 P Eudicot Erythrina Gleditsia suberosa ferox Roxb. Desf. 42 28 2 2 P P Eudicot Eudicot Gleditsia sinensis Lam. 28 2 P Eudicot Laburnum anagyroides Med. 48 4 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Hiramath et al., 2001 Ohri, 1996 Ohri and Kumar, 1986 Ohri and Kumar, 1986 Olszewska and Osiecka, 1984 P Eudicot 36 Eudicot Eudicot Laburnum alpinum Mimosa pudica Mimosa invisa Prosopis Prosopis Prosopis Prosopis Prosopis Prosopis Prosopis Prosopis Prosopis flexusa alba lampa pallida glandulosa siliquas juliflora juliflora chilensis Prosopis chilensis Prosopis Robinia cineraria pseudoacacia Senna alexandrina Sesbania rostrata Tamarindus Acacia Acacia Acacia Acacia Acacia Acacia Acacia Acacia Acacia Acacia Acacia indica modesta catechu seyal albida sieberana nubica implexa mearnsii sophorae polycantha heterophylla Acacia falcata Acacia bivenosa Acacia raddiana Brecht Persl & 48 4 52 4 Mart 26 2 DC. Griseb. Willd. Willd. Torr. (Willd.) (Swartz) DC. DC. (Molina) Stuntz. (Molina) Stuntz. (L.) Druce L. 28 28 28 28 28 28 28 52 28 56 Bremek. Oberm L. Wall. Willd. (Del.) (Del.) DC. Benth. Benth. Willd. Benth. Willd. P Eudicot P Eudicot B-P Eudicot 2 2 2 2 2 2 2 4 2 Olszewska and Osiecka, 1984 Bennett et al., 1998 Bennett et al., 1998 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Ohri, 1996 Bukhari, 1997 P P P P P P P P P Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot 4 Bukhari, 1997 P Eudicot Ohri, 2002 P Olszewska and P Osiecka, 1984 Al-Turki et al., (2000) Eudicot Eudicot Arumuganathan and Earle, 1991 Ohri, 2002 Ohri, 1996 Ohri, 1996 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Bukhari, 1997 Marie and Brown, 1993 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 - Eudicot P P P P P P P P P P P - Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot P Eudicot P Eudicot P Eudicot 52 20 2 28 2 & 12 2 26 26 26 104 26 26 56 26 26 26 52 26 2 2 2 8 2 2 4 2 2 2 4 2 26 2 DC. 26 2 Savi 104 8 37 Acacia caffra 26 2 Bukhari, 1997 P Eudicot senegal (Thunb.) Willd. Willd. Acacia 26 2 P Eudicot Acacia tortilis Hayne 78 6 P Eudicot Acacia crassa Pedley 26 2 P Eudicot Acacia orites Pedley 26 2 P Eudicot Acacia raddiana 78 6 P Eudicot Acacia nilotica (Savi.) Brenan Delile Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Bukhari, 1997 26 2 P Eudicot Acacia salicina Lindl. 26 2 P Eudicot Acacia gerrardi Benth. 52 4 P Eudicot Acacia mellifera 26 2 P Eudicot Acacia tortilis 52 4 Bukhari, 1997 P Eudicot Acacia tortilis 104 8 Bukhari, 1997 P Eudicot Acacia mangium (Vahal) Benth. (Forssk.) Hayne (Forssk.) Hayne Willd. Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Bukhari, 1997 26 2 P Eudicot Acacia dealbata Link. 26 2 P Eudicot Acacia dealbata Link. 39 3 P Eudicot Acacia dealbata Link. 52 4 P Eudicot Acacia ligulata A.Cunn. 26 2 P Eudicot Acacia suberosa A.Cunn. 26 2 P Eudicot Acacia victoriae Benth. 26 2 P Eudicot Acacia falciformis DC. 26 2 P Eudicot Acacia irrorata Sieber 26 2 P Eudicot Acacia melanoxylon R.Br. 26 2 P Eudicot Acacia farnesiana Willd. 52 4 Blakesley et al., 2002 Blakesley et al., 2002 Blakesley et al., 2002 Blakesley et al., 2002 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 P Eudicot 38 Acacia bancrofti Maiden 26 2 P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot 4 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Bukhari, 1997 Acacia pycnantha Benth. 26 2 Acacia leucophloea Willd. 52 4 Acacia hylonoma L.Pedley 26 2 Acacia maidenii F.Muell. 26 2 Acacia concurrens Pedley 26 2 Acacia nilotica 52 Acacia Acacia drepanolobiu m nilotica Acacia verek Acacia longispicata (L.) Willd. ex Del. Harm. ex Sios. (L.) Willd. ex Del. Guill. & Perr. Benth. P Eudicot 52 4 Bukhari, 1997 P Eudicot 104 8 Bukhari, 1997 P Eudicot 26 2 P Eudicot 26 2 P Eudicot F.Muell. 26 2 P Eudicot A.Cunn. 26 2 P Eudicot Acacia auriculaeform is podalyriifolia A.Cunn. 26 2 P Eudicot Acacia leiocalyx (Domin) Pedley 26 2 P Eudicot Acacia simsii 26 2 P Eudicot Acacia greggii 26 2 P Eudicot Acacia fimbriata 26 2 P Eudicot Acacia leptocarpa 26 2 P Eudicot Acacia holosericea 52 4 P Eudicot Acacia crassicarpa 26 2 P Eudicot Acacia aulacocarpa 26 2 P Eudicot Acacia iteaphylla 26 2 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Mukherjee and Sharma, 1995 Acacia cincinnata Acacia P Eudicot A.Gray, P.L.Wright A.Cunn. ex G.Don A.Cunn. ex Benth. A.Cunn. ex G.Don A.Cunn. ex Benth. A.Cunn. ex Benth. F.Muell. ex Benth. 39 Cassia Cassia Cassia Cassia Cassia Cassia Cassia Cassia tora absus fistula biflora siamea glauca auriculata occidentalis L. L. L. L. Lam. Lam. L. L. 26 28 28 28 28 28 28 28 2 2 2 2 2 2 2 2 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 Ohri et al., 1986 A A P P P P P A Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Eudicot Cassia excelsa Schrad. 28 2 Ohri et al., 1986 P Eudicot Cassia australis ex 28 2 Ohri et al., 1986 P Eudicot Cassia angustifolia Reinw. Steud. Vahl. 28 4 - Eudicot Leucaena retusa Benth. 56 2 P Eudicot Leucaena lanceolata S.Watson 56 2 P Eudicot Leucaena greggii S.Watson 56 2 P Eudicot Leucaena confertiflora Zárate 112 4 P Eudicot Leucaena shannonii J.D.Smith 52 2 P Eudicot Leucaena trichodes 52 2 P Eudicot Leucaena diversifolia 56 2 P Eudicot Leucaena diversifolia 104 4 P Eudicot Leucaena pulverulenta 56 2 P Eudicot Leucaena esculenta 56 2 P Eudicot Leucaena esculenta 112 4 Palomino et al., P 1995 Eudicot Lonchocarpus xuul (C.E.Hughes ) C.E.Hughes (Schltdl.) Benth. (Schltdl.) Benth. (Schltdl.) Benth. (Mociño & Sessé ex A.DC.) Benth. (Mociño & Sessé ex A.DC.) Benth. Lundell Bahl and Srivastava, 1987 Hartman et al., 2000 Palomino et al., 1995 Hartman et al., 2000 Palomino et al., 1995 Hartman et al., 2000 Hartman et al., 2000 Palomino et al., 1995 Hartman et al., 2000 Hartman et al., 2000 Palomino et al., 1995 22 2 Eudicot Lonchocarpus mutans M.Sousa 22 2 Palomino and P Sousa, 2000 Palomino and P Sousa, 2000 40 Eudicot Lonchocarpus rugosus Benth. 22 2 Lonchocarpus obovatus Benth. 22 2 Lonchocarpus punctatus Kunth 22 2 Lonchocarpus hintonii Sandw. 22 2 Lonchocarpus morenoi M.Sousa 22 2 Lonchocarpus chiangii M.Sousa 22 2 Lonchocarpus cruentus Lundell 22 2 Lonchocarpus hughesii M.Sousa 22 2 Lonchocarpus caudatus Pittier 22 2 Lonchocarpus epigaeus M.Sousa 22 2 Lonchocarpus castilloi Standl. 22 2 Lonchocarpus hermannii M.Sousa 22 2 Lonchocarpus parviflorus Benth. 22 2 Lonchocarpus agyrotrichus Harms 22 2 Lonchocarpus andrieuxii M.Sousa 22 2 Lonchocarpus eriophyllus Benth. 22 2 Lonchocarpus macrocarpus Benth. 22 2 Lonchocarpus lanceolatus Benth. 22 2 Lonchocarpus emarginatus Pittier 22 2 Lonchocarpus spectabilis Hermann 22 2 Lonchocarpus constrictus Pittier 22 2 Lonchocarpus hidalgensis Lundell 22 2 Lonchocarpus schubertiae M.Sousa 22 2 41 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 Palomino and Sousa, 2000 P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot P Eudicot Lonchocarpus atropurpureus Benth. 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus guatemalensis Benth. 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus phaseolifolius Benth. 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus eriocarinalis Micheli 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus salvadorensis Pittier 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus luteomaculatu Pittier 22 2 Palomino and P Eudicot s Sousa, 2000 Lonchocarpus santarosanus Donn. Sm. 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus minimiflorus Donn. Sm. 22 2 Palomino and P Eudicot Sousa, 2000 Lonchocarpus chavelasii M.Sousa, 22 2 Palomino and P Eudicot ined. Sousa, 2000 Lonchocarpus martinezii M.Sousa, 22 2 Palomino and P Eudicot ined. Sousa, 2000 Lonchocarpus molinae Standl. & 22 2 Palomino and P Eudicot L.O.Wms. Sousa, 2000 Lonchocarpus torresiorum M.Sousa, 22 2 Palomino and P Eudicot ined. Sousa, 2000 Lonchocarpus schiedeanus (Schldl.) 22 2 Palomino and P Eudicot Harms Sousa, 2000 Lonchocarpus angusticarpus M.Sousa, 22 2 Palomino and P Eudicot ined. Sousa, 2000 Lonchocarpus balsensis M.Sousa & 22 2 Palomino and P Eudicot J.C.Soto Sousa, 2000 Lonchocarpus acuminatus (Schldl.) 22 2 Palomino and P Eudicot M.Sousa Sousa, 2000 Lonchocarpus sanctuari Standl. & 22 2 Palomino and P Eudicot L.O.Wms. Sousa, 2000 Lonchocarpus peninsularis (J.D. Sm.) 22 2 Palomino and P Eudicot Pittier Sousa, 2000 Lonchocarpus huetamoensis M.Sousa & 22 2 Palomino and P Eudicot J.C.Soto Sousa, 2000 Lonchocarpus phlebophyllus Standl. & 22 2 Palomino and P Eudicot Steyerm. Sousa, 2000 Lonchocarpus longipeduncul M.Sousa & 22 2 Palomino and P Eudicot atus J.C.Soto Sousa, 2000 Data were from the Plant DNA C-values Database query results (http://www.rbdkew.org.uk/cval/homepage.html). 42 CHAPTER FIVE 5.0 CONCLUSION 5.1 CHROMOSOME STUDIES IN TREE LEGUME It was observed that most sampled tree legume have their chromosome number n = 13. The ploidy level ranges from 2 to 8. It was observed that there is constant chromosome number of 22 in the entire Lonchocarpus trees assessed with their ploidy level at 2. Cassia and Leucaena show similaties in their chromosome number though with little variation in their ploidy levels. It was noted that they both have their chromosome number n = 14 with very few of them having their chromosome number n = 13. The few species of Cassia, with n = 13, show similarity with the genus Acacia that have haploid chromosome numbers of n = 13. It was observed also that Prosopis and Cassia has the same chromosome number of 28. Likewise Abrus, Cajanus and Lonchocarpus have same haploid number of chromosomes as n = 22. The similarities or closeness observed in theses tree legume indicates the closeness of these genera. Between the genera Cassia with the highest and Astragalus with the lowest chromosome numbers, there are intermediate genera to bridge the gap of relatedness among the tree legume. 5.2 PLOIDY LEVEL IN TREE LEGUME It was also observed within the genera of the tree legume that as there are variations within same genus of tree legumes, there is constancy in the chromosome number within another where there are no multiples of single basic number. Some genera with different ploidy level in their chromosomes include Mimosa, Leucaena, Acacia, Prosopis and Cassia. These genera are euploid. Some genera with constancy in their chromosome ploidy level include Cajanus, Lonchocarpus and Dalbergia. The genera Astragalus, Robinia, Senna, Sesbania, Erythrina, 43 Gleditsia, Laburnum, Abrus, Albizzia, Alhagi, and Bauhinia are under-represented to justify the constancy or differences of the chromosome numbers and ploidy levels. From the Table 1, only one species in the genera Cassia and Prosopis show different ploidy level which consist entirely of multiples of their single basic number. It was also observed that in the majority of literatures consulted for the Table 1, only the chromosome number and the ploidy level of the tree legumes were recorded. This could indicate that the type of chromosomes in relation to the position of the centromere has no major documentation. Other data that were not commonly documented for these tree legumes include the relative sizes of their chromosomes and the number and positions of their satellite (if any). Also not listed is the degree and distribution of heterochromatic regions of these chromosomes. 44 REFERENCE Adams K.L. & Wendel J.F. 2005. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 8 135-41. Agnieszka M, Hitoshi M, Keiichi O. 2006. The origin of Darwin hybrid tulips analyzed by flow cytometry, karyotype analyses and genomic in situ hybridization. Euphytica, 151: 279290. Ahn, S., and Tanksley, S.D. 1993. Comparative linkage maps of rice and maize genomes. Proc. Natl. Acad. Sci. USA 90, 7980–7984. AL-Turki Turki A., Shafik A. Filfilan & Syed F. Mehmood. 2000. A cytological study of flowering plants from Saudi Arabia. Willdenowia 30, 339-358. Arumuganathan K, Earle ED. 1991. Nuclear DNA content of some important plantspecies. Plant Molecular Biology Reporter 9: 208-218. Ayşe Nihal Gömürgen and Haşim Altinözlü. 2005. Chromosome Number and Karyotype Analysis of Kalidiopsis Wagenitzii Aellen, Pak. J. Bot., 37(2): 307-311. Badr A. and A. Gasim, 1992. chromosomal studies on some plants in the floral of Medina region. J. K. A. U. Sci., Vol 4, pp. 23-25 Bahl JR, Srivastava HK. 1987. Nuclear DNA variations in Withania somnifera Dun. and Cassia angustifolia Vahl. Nucleus 30: 99-100. Bennett MD, Leitch IJ, Hanson L. 1998. DNA amounts in two samples of angiosperm weeds. Annals of Botany 82 (Supplement A): 121-134. 45 Bennett MD, Smith JB. 1976. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London Series B - Biological Sciences 274:227-274. Bir, S. S. & Kumari, S. 1977: Evolutionary status of Leguminosae from Pachmarhi, central India. Nucleus 20: 94-98. Blakesley D, Allen A, Pellny TK, Roberts AV. 2002. Natural and induced polyploidy in Acacia dealbata Link. and Acacia mangium Willd. Annals of Botany 90: 391-398. Borgen, L. 1980: Chromosome numbers of Macaronesian flowering plants III. Bot. Macaron 7: 67-76.Brutovska R, Kusnirikova P, Bogyiova E, Cellarova E (2000). Karyotype analysis of Hypericum perforatum L. Biol. Plant 43(1):133-136. Brewbaker, J.L. 1986. Leguminous trees and shrubs for Southeast Asia and the South Pacific. In: Blair, G.J., Ivory, D.A. and Evans, T.R. (eds), Forages in Southeast Asia and South Pacific Agriculture. Proceedings of an international workshop held at Cisarua, Indonesia. ACIAR Proceedings Series No. 12, Canberra, pp. 43-50. Brubaker, C.L., Paterson, A.H., and Wendel, J.F. 1999. Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42, 184–203. Bukhari YM. 1997. Nuclear DNA amounts in Acacia and Prosopis (Mimosaceae) andtheir evolutionary implications. Hereditas 126: 45-51. Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews, Genetics. 6, 836-46. 46 Commonwealth Agricultural Bureaux .1947. International Advances in Grassland Husbandry and Fodder Production. Second Symposium, CAB Farnham Royal, UK. Darlington C.D. 1939. Evolution of genetic systems. Cambridge University Press. 2nd ed, revised and enlarged, 1958. Oliver & Boyd, Edinburgh. Dolja Pavlova and Anita Tosheva. 2004. Notes on karyomorphology of Melilotus officinalis populations in Bulgaria. Caryologia Vol. 57, no. 2: 151-157. Dougall, H.W. and Bogdan, A.V. 1958. Browse plants in Kenya. East African Agricultural Journal, 236-246. Doyle JJ, Luckow MA .2003. The rest of the iceberg: legume diversity and evolution in a phylogenetic context. Plant Physiol 131: 900–910 Eckholm, E. 1975. The other energy crisis: firewood. Worldwatch Paper No. 1. Worldwatch Institute, Washington, DC. Edgar B.A. & Orr-Weaver T.L. 2001. Endoreduplication cell cycles: more for less. Cell 105, 297-306. Engelhardt H .1922. Die alttertiare flora von messel bei darmstadt. In Abhandlungen der Hessischen Geologischen Landesanstalt zu Darmstadt, Band VII, Heft 4. Hessischer Staatsverlag, Darmstadt, Germany, pp 21–23. Federov A., ed. 1974. Chromosome numbers of flowering plants. Reprint by Otto Koeltz Science Publishers, Koenigstein, Germany. 47 Franzen JL, Gingerich PD, Habersetzer J, Hurum JH, von Koenigswald W, Smith BH (2009) Complete primate skeleton fromtheMiddle Eocene of Messel in Germany: morphology and paleobiology. PLoS One 4: e5723. Fuchs J, Strehl S, Brandes A et al. (1998a) Molecular-cytogenetic characterization of the Vicia faba genome-heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res 6: 219-230. George, S. M. & Bhavandan, K. V. 1993: Cytological studies in some species of Cassia from south India. J. Cytol. Genet. 28: 1-5. Gill, L. S. and A. O. Obembe, 1991. Chromosome Studies in Some Trees and Shrubs from Southern Nigeria. Willdenowia, Bd. 21, H. 1/2, pp. 233-238 Gill, L. S. & Husaini, S. W. 1986: Cytological observations in Leguminosae from southern Nigeria. Willdenowia 15: 521-527. Goldblatt P. 1981a. Cytology and the phylogeny of Leguminosae. Pages 427-463 in R. M. Polhill and P. H. Raven, eds. Advances in legume systematics, Part 2. Royal Botanic Gardens, Kew. Goldblatt P. 1981b. Chromosome numbers in legumes. II. Ann. Mo. Bot. Gard. 68:551-557. Goldblatt P. ed. 1981c. Index to plant chromosome numbers. 1975 - 1978. Monogr. Syst. Bot. 5:1-259. Goldblatt P. 1984. Index to plant chromosome numbers. 1979-1981. Monogr. Syst. Bot.8:1-194. Goldblatt P. 1988. Index to plant chromosome numbers. 1984-1985. Monogr. Syst. Bot. 23:1122. 48 Goldblatt P. 1990. Index to plant chromosome numbers. 1986-1987. Monogr. Syst. Bot. 30:1243. Goldblatt P. 1991. Index to plant chromosome numbers. 1988-1989. Monogr. Syst. Bot. 40:1238. Goldblatt P. 1994. Index to plant chromosome numbers. 1990-1991. Monogr. Syst. Bot. 51:1267. Goldblatt P. 1996. Index to plant chromosome numbers. 1992-1993. Monogr. Syst. Bot. 58:1276. Goldblatt P. 1998. Index to plant chromosome numbers. 1994-1995. Monogr. Syst. Bot. 69:1 208. Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872-877. Gregory T.R. & Mable B.K. 2005. Polyploidy in animals. In The Evolution of the genome Gregory T.R. (ed). Elsevier, San Diego. p427-517 Guerra, M. 1983. O uso do Giemsa em citogenética vegetal – comparação entre a coloração simples e o bandeamento. Ciênc. e Cult. 35: 190-193. Guilhem Mansion and Louis Zeltne. 2004. Phylogenetic relationships within the new world endemic zeltnera (gentianaceae- chironiinae) inferred from molecular and karyological data. American Journal of Botany 91(12): 2069-2086. 49 Gustashaw K.M. 1991. Chromosome stains. In The ACT Cytogenetics Laboratory Manual 2nd ed, ed. M.J. Barch. The Association of Cytogenetic Technologists, Raven Press, New York. Halverson, K., S. B. Heard, J. D. Nason, and J. O. Stireman III. 2008. Origin, distribution, and local co-occurence of polyploid cytotypes in Solidago altissima (Asteraceae). American Journal of Botany 95:50–58 Hartman TPV, Jones J, Blackhall NW, Power JB, Cocking EC, Davey MR. 2000.Cytogenetics, molecular cytogenetics and genome size in Leucaena (Leguminosae,Mimosideae). In: Guttenberger H, Borzan Z, Schlarbaum SE, Hartman TPV eds.Cytogenetic studies of forest trees and shrubs - review, present status, andoutlook on the future. Zvolen, Slovakia: Arbora Publishers, 57-70. Hill R.D., Myers W.M. 1945. A schedule including cold treatment to facilitate somatic chromosome counts in certain forage grasses. Stain Tech. 20:89–92. Ijdo J.W. et al 1991. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proceedings of the National Academy of Sciences 88: 9051–5. Jones, R.J. (1986) Overcoming the Leucaena toxicity problem to realise the potential of Leucaena Australian Institute of Agricultural Science, Queensland Branch, Bulletin No. 289, pp. 4-9. Kanda H. and Okada H. 1993. Chromosome study on the submerged moss collected from Antarctic lakes. Proc. NIPR Symp. Polar Biol., 6, 121-125. 50 Kazuo O, Tanguy J, Hiroshi T (2000). The Karyotype Analysis of Somatic Chromosomes in Amfwrella trichopoda (Amborellaceae). J. Plant Res, 113: 281-283. Kazuo NW, Matilde O (1993). An alternative pretreatment method for mitotic chromosomes observation in potatoes. Am. Potato J. 70: 543-548. Khatoon, S. 1991: Polyploidy in the flora of Pakistan – an analytical study. Ph. D. thesis, University of Karachi. King R.C., Stansfield W.D. and Mulligan P.K. 2006. A dictionary of genetics. 7th ed, Oxford U.P Oxford & N.Y. p242 Kron, P., J. Suda, and B. C. Husband. 2007. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology and Systematics 38:847–876. Kumari, S. & Bir, S. S. 1990: Karyomorphological evolution in Papilionaceae. J. Cytol. Genet. 25: 173-219. Lagercrantz, U., and Lydiate, D. (1996). Comparative genome mapping in Brassica. Genetics 144, 1903–1910. Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594. Ledingham, G. F. 1960: Chromosome numbers in Astragalus and Oxytropis. Canad. J. Genet. Cytol. 2: 119-128. Lessani, H. & Chariat-Panahi, S. 1979: Reports. – [In: Löve, Á. (ed.), IOPB chromosome number reports LXV]. – Taxon 28: 635-636 51 Levin, D. A. 2002. The role of chromosomal change in plant evolution. Oxford University Press, New York Levitsky G.A. 1924. The material basis of heredity. State Publication Office of the Ukraine, Kiev. Levitsky G.A. 1931. The morphology of chromosomes. Bull. Applied Bot. Genet. Plant Breed. 27, 19-174. Lewis G, Schrire B, Mackind B, Lock M (2005) Legumes of the World. Royal Botanic Gardens, Kew, UK Li J, Chen SM and Chen FD (2008). Karyotype and meiotic analyses of six species in the subtribe Chrysantheminae. Euphytica, 164: 293-301. Li, M.X. 1982. Introducing a good stain used for nuclei and chromosome. Bull. Biol., 5: 53. Lilly M.A. & Duronio R.J. (2005). "New insights into cell cycle control from the Drosophila endocycle.". Oncogene 24 (17): 2765–75. Lisa G. Shaffer and Niels Tommerup, ed (2005). ISCN 2005: An International System for Human Cytogenetic Nomenclature. Switzerland: S. Karger AG. Lv Lingling, Guangming Sun, Jianghui Xie, Xiaoping Zang, Yulin Hu and Jun Duan. 2009. Determination of chromosomal ploidy in Agave ssp. African Journal of Biotechnology Vol. 8 (20), pp. 5248-5252. 52 Matzke, M.A., and Matzke, A.J.M. (1998). Polyploidy and transposons. Trends Ecol. Evol. 13, 241. Marie D, Brown SC. 1993. A cytometric exercise in plant DNA histograms, with 2Cvalues for 70 species. Biological Cell 78: 41-51. McKell, C.M. (1980) Multiple use of fodder trees and shrubs - a worldwide perspective. In: Le Houerou, H.N. (ed.), Browse in Africa. ILCA, Addis Ababa, Ethiopia, pp. 141-150. Medina, D.M. and Conagin, C.H.T.M. (1964). Técnica Citológica. Publicação No. 2610. Instituto Agrônomico, Campinas. Menancio-Hautea D, Fatokum CA, Kumar L, Danesh D, Young ND (1993). Comparative genome analysis of mungbean (Vigna radiata (L.) Wilczek) and cowpea (V. ungui cul a t a (L.) Walpers) using RFLP mapping data. Theor Appl Genet 86: 797–810. Meyer, J. R. 1945. Prefixing with paradichlorobenzene to facilitate chromosome study. Stain Techn. 20: 121-125. Moog, F.A. (1985) Forages in integrated food cropping systems. In: Blair, G.J., Ivory, D.A. and Evans, T.R. (eds), Forages in Southeast Asian and South Pacific Agriculture. Proceedings of an international workshop held at Cisarua, Indonesia. ACIAR Proceedings Series No. 12, Canberra, pp. 152-156. Morphology". Www.wikipedia.com. Retrieved 05-10-2010. Mukherjee S, Sharma AK. 1995. In situ nuclear DNA variation in Australian species of Acacia. Cytobios 83: 59-64. 53 Murray, B. G., and A. G. Young. 2001. Widespread chromosome variation in the endangered grassland forb Rutidosis leptorrhynchoides F. Muell. (Asteraceae: Gnaphalieae). Annals of Botany 87:83–90 Nagl W. 1978. Endopolyploidy and polyteny in differentiation and evolution: towards an understanding of quantitative and qualitative variation of nuclear DNA in ontogeny and phylogeny. Elsevier, New York. Nankui T, Paul WB (2003). Observations on interspecific compatibility and meiotic chromosome behavior of Capsicum buforum and C. lanceolatum. Genet. Res. Crop Evol. 50: 193-199. NAS (1980) Firewood Crops Shrub and Tree Species for Energy Production Volume I. National Academy Press, Washington DC, 235 pp. NAS (1983) Firewood Crops Shrub and Tree Species for Energy Production Volume II. National Academy Press, Washington DC, 92 pp. Nathewet P, Tomohiro Y, Kazumi S, Shin T, Nobuyuki O (2007). Chromosome observation method at metaphase and pro-metaphase stages in diploid and octoploid strawberries. Scientia Hortic. 114: 133-137. O’Mara J.G. 1939. Observations on the immediate effect of colchicine. J. Hered. 30:35–37. Oginuma Kazuo and Hiroshi Tobe .1995. Karyomorphology of some Moraceae and Cecropiaceae (Urticales) Journal of Plant Research 108(3), 313-326 54 Ohmido Nobuko, Shusei Sato, Satoshi Tabata & Kiichi Fuku. 2007. Chromosome maps of legumes. Chromosome Research 15:97–103 Ohri D. 1996. Genome size and polyploidy variation in the tropical hardwood genus Terminalia (Combretaceae). Plant Systematics and Evolution 200: 225-232. Ohri D. 2002. Genome size variation in some tropical hardwoods. BiologiaPlantarum 45: 455457. Ohri D, Kumar A, Pal M. 1986. Correlations between 2C DNA values and habit in Cassia (Leguminosae: Caesalpinioideae). Plant Systematics and Evolution 153: 223-227. Ohri D, Singh PS. 2002. Karyotypic and genome size variation in Cajanus cajan(L.) Millsp. (pigeonpea) and some wild relatives. Genetic Resources and Crop Evolution 49: 1-10. Olszewska MJ, Osiecka R. 1984. The relationship between 2C DNA content,systematic position, and the level of nuclear DNA endoreplication duringdifferentiation of root parenchyma in some dicotyledonous shrubs and trees -comparison with herbaceous species. Biochemie Und Physiologie Der Pflanzen 179:641-657. Östergren G. and Heneen W. K. 2009. A squash technique for chromosome morphological studies. Hereditas Volume 48 Issue 1-2, Pages 332 - 341 Palomino G, Romo G, Zárate S. 1995. Chromosome numbers and DNA content in sometaxa of Leucaena (Fabaceae Mimosoideae). Cytologia 60: 31-37. 55 Palomino G, Sousa SM. 2000. Variation of nuclear DNA content in the biflorus species of Lonchocarpus (Leguminosae). Annals of Botany 85: 69-76. Polhill RM, Raven PH, eds (1981) Advances in Legume Systematics, Part 1. Royal Botanic Gardens, Kew, UK Polymorphism: when two or more clearly different phenotypes exist in the same interbreeding population of a species. Ford E.B. 1975. Ecological genetics, 4th ed. Ramon RC, Manuel UA (2004). Karyotype analysis of Eucinostomus argenteus, E. gula, E. harengulus, and Eugerres plumieri (Teleostei, Gerreidae) from Florida and Puerto Rico. Environ. Biol. Fishes. 67: 269-276. Reinisch, A.R., Dong, J.-M., Brubaker, C., Stelly, D., Wendel, J., and Paterson, A.H. (1994). An RFLP map of cotton (Gossypium hirsutum and G. barbadense): Chromosome organization and evolution in a disomic polyploid genome. Genetics 138, 829–847. Reynolds, C. and Atta-Krah, A.N. (1986) Alley farming with livestock. In: Kang, B.T. and Reynolds, L. (eds), Alley Farming in the Humid and Subhumid Tropics. Proceedings of an international workshop held at Ibadan, Nigeria pp. 27-36. Robinson, P.J. (1985) Trees as fodder crops. In: Cannell, M.G.R. and Jackson, J.E. (eds), Attributes of Trees as Crop Plants. Institute of Terrestrial Ecology, Huntingdon, UK, pp. 281-300. Ross C. Gutteridge and H. Max Shelton (ed), Forage Tree Legumes in Tropical Agriculture,1994 , CAB Intemational, Wallinford, Oxon, UK. ISBN 0 958567719 56 Schmuck A., Kostoff D. 1939. Bromoacenaphthaline and bromonaphthaline as agents inducing chromosome doubling in rye and wheat. C.R. (Doklady) Acad. Sci URSS. 23:263–266. Severns Paul M. and Aaron Liston. 2008. Intraspecific Chromosome Number Variation: a Neglected Threat to the Conservation of Rare Plants. Conservation Biology, 22(6), 1641– 1647. Skerman, P.J. (1977) Tropical Forage Legumes. FAO Plant Production and Protection Series No. 2. FAO, Rome, 609 pp. Soltis, D. E., P. S. Soltis, D. W. Schemske, J. F. Hancock, J. N. Thompson, B. C. Husband, and W. S. Judd. 2007. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56:13–30. Song, K., Lu, P., Tang, K., and Osborn, T.C. (1995a). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92, 7719–7723. Song, K., Slocum, M.K., and Osborn, T.C. (1995b). Molecular marker analysis of genes encoding morphological variation in Brassica rapa (syn. campestris). Theor. Appl. Genet. 90, 1–10. Stalfelt, M. G. 1950. The effect of oxyquinoline on protoplasmic viscosity. - Appendix to the paper of Tjio and Levan (1950). Stiefkens Laura B., Gabriel Bernardello, and Gregory J. Anderson. 2001. The Somatic Chromosomes of Sophora fernandeziana (Fabaceae), an Endemic Tree from Robinson Crusoe Island. Pacific Science 55(1):71-75 57 Stebbins, G.L. 1940. The significance of polyploidy in plant evolution. The American Naturalist 74, 54–66. Stebbins G.L. 1950. Variation and evolution in plants. Chapter XII: The Karyotype. Columbia University Press N.Y. Stebbins G.L. 1970. Chromosomal evolution in flowering plants. Arnold, London. Stebbins G.L. 1971. Chromosomal evolution in higher plants. Arnold, London. p85-6 Stebbins, G. Ledley, Jr. 1972. Chromosomal evolution in higher plants. Nelson, London. p18 Steele, K.W. and Vallis, I. (1988) The nitrogen cycle in pastures. In: Wilson, J.R. (ed.), Advances in Nitrogen Cycling in Agricultural Ecosystems. CAB International, Wallingford, UK, pp. 274-291. Steven B. Cannon, Gregory D. May, and Scott A. Jackson. 2009. Three Sequenced Legume Genomes and Many Crop Species: Rich Opportunities for Translational Genomics. Plant Physiology, Vol. 151, pp. 970–977. Tabur S. and Oney S. 2009, Effect of artificial fertilizers on mitotic index and chromosome behaviour in Vicia hybrida l., J. Agric. Res. 47(1) Tacio, H.D., Watson, H.R. and Laquihon, W.A. (1987) Nitrogen fixing trees as multipurpose species for soil conservation. In: Multipurpose Tree Species for Small-farm Use. Proceedings of an international workshop held in Pattaya, Thailand, pp. 114-121. Tian-Li Wang, Christine Maierhofer, Michael R. Speicher, Christoph Lengauer, Bert Vogelstein, Kenneth W. Kinzler , and Victor E. Velculescu. 2000. Digital karyotyping - Proceedings of the National Academy of Sciences 58 Tjio, J. H. and Levan, A. 1950. The use of oxyquinoline in chromosome analysis. Anales Est. Exp. Aula Dei 2: 2144. Weeden NF, Muehlbauer FJ, Ladizinsky G (1992) Extensive conservation of linkage relationships between pea and lentil geneticmaps. JHered 83: 123–129 White M.J.D. 1973a. The chromosomes. 6th ed, Chapman & Hall, London. P28 White M.J.D. 1973b. Animal cytology and evolution. 3rd ed, Cambridge University Press. White M.J.D. 1973c. The chromosomes. 6th ed, Chapman & Hall, London. p45 White M.J.D. 1973d. The chromosomes. Chapman & Hall, London. p169 Williams, RJ. 1983. Tropical Legumes. In: McIvor, J.G. and Bray, R.A. (eds), Genetic Resources of Forage Plants. CSIRO, Melbourne, pp. 17-37. Yeh, M. S., Yuasa, H. and Maekawa, F. 1986: Chromosome numbers in the Leguminosae. Sci. Rep. Res. Inst. Evol. Biol. 3: 57-71. Young Nevin Dale, Joann Mudge and TH Noel Ellis. 2003. Legume genomes: more than peas in a pod. Current Opinion in Plant Biology, 6:199–204 Zhao, X., Si, Y., Hanson, R., Price, H.J., Stelly, D., Wendel, J., and Paterson, A.H. (1998). Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 8, 479–492. Zoleka Mhinana, Buyisile Mayekiso and Michael L. Magwa. 2010. Anatomy and morphology of Nicotiana glauca with regard to its crystals characterization. African Journal of Plant Science Vol. 4(6), pp. 172-178. 59