Supplementary Figure 7 - Springer Static Content Server
advertisement

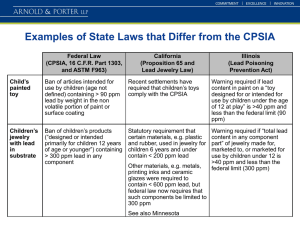
Supplementary Table and Figures Supplementary Table 1 Expected HMBC correlation patterns for hop bitter acid acyl side chain signals Compound Me group 2 (1H signal multiplicity) J & 3J HMBC to C Humolone, 4’ (d) 2’ (CH2); 3’ (CH); 5’ (CH3) Lupolone 5’ (d) 2’ (CH2); 3’ (CH); 4’ (CH3) Cohumulone, 3’ (d) 1’ (C=O); 2’ (CH2); 4’ (CH3) Colupolone 4’ (d) 1’ (C=O); 2’ (CH2); 3’ (CH3) Adhumulone, 4’ (t) 2’ (CH); 3’(CH2) Adlupolone 5’ (d) 1’ (C=O); 2’ (CH); 3’(CH2) Supplementary Figure 1 Traces parallel to the 1H axis of different HMBC spectra of HHS resin under variation of number of scans and number of increments. Normalized signal intensity of the minor cross peak at 1H/13C of 3.24/108.5 ppm assigned to C-7 of the enol tautomer of lupulones is displayed (indicated by an arrow). HMBC spectra were recorded at three different acquisition parameters: (a) 2 scans and 256 increments; (b) 8scans and 256 increments and (c) 16 scans and 192 increments for total acquisition times of 28, 89 and 146 min, respectively Supplementary Figure 2 HMBC spectrum of HHE hop resin acquired in CD3OD. As outlined in the text, the spectrum can be roughly divided into 4 main regions along the F2 proton dimension labelled as: I: 1H 5.5 - 8.5 ppm; II: 1H 4.0 - 5.5 ppm; III: 1H 1.8 - 4.0 and IV: 1H 0.0 - 1.8 ppm Supplementary Figure 3 (a) General structure of α-acids and iso α-acids showing the change in the hybridization status of C-5 from sp2 to sp3 and its associated 13 C upfield chemcal shift (in ppm). Arrows represent HMBC correlations that were readily observed in reference HMBC spectra of α-acids (b) Partial HMBC spectrum of aged solution of hops α acids showing different marker cross peaks for α-bitter acids and their isomerization products labelled as follows: 1, H2-12 C-6 of α-bitter acids (humulones) 2, H2-7 C-4 of cis-isohumulones 3, H2-7 C-5 of cis-isohumulones 4, H2-7 C-5 of trans-isohumulones 5, H2-7 C-5 of dihydro trans-isohumulones Other signals could not be assigned due to lack of literature NMR data Supplementary Figure 4 Flowchart showing the path of data extraction from a HMBC spectrum to principal component analysis. Step (1): acquisition of HMBC spectra of hop samples using Agilent (Varian) (CHEMPACK 5.1) spectrometer software. Step (2): Integration of HMBC cross peaks using ACD/NMR Manager lab version 10.0 software to reduce the complex HMBC profile into pixels (i.e. constant small area2D buckets) that cover the 1H and 13C dimensional grid. Step (3): Integration volumes are then plotted against different pixels to generate a comprehensive 2D NMR chemomatrix. Step (4): The generated chemomatrix is fed into statistic software R (2.9.2) using Bioconductor package pcaMethods together with customwritten procedures (Supplementary Word File, or Download from ipb-halle.de) to produce scores and loading plots for classification of samples. 1 2 TPE HPE ○ 40 A HTU 0 EHM HHT ENB -20 HHE -40 PC2 (18.1%) 20 HSE CZAG HHS ▪ Pixel position Intensity H0.74/C208.3 0,741 H0.74/C208.3 208,3085 0,155 0,741 0,145 ATPE ATHM HHM ∆ -100 -50 PC1 (67.5%) 4 0 50 3 208,3085 0,1 Supplementary Figure 5 PCA scoring plots of PC1 and PC2 scores using different pixel sizes in both F1 and F2 dimensions (F1xF2): a, (85x128); b, (64x256); c, (85x256) and d, (128x256). HMBC cross peaks intensities were recorded in the F1 range ( -6.0-231) ppm and F2 range (0.4–8.5) ppm. Triplicates of both CZAG and HSE cultivars are displayed individually as an indicator of extraction & spectral reproducibility, respectively. Supplementary Figure 6 Loading plot for PC1 components of the region from 1H 0.4-3.6 ppm and 13C 0-230 ppm showing cross peaks contributing to PC1 variance. Signals with positive contribution to PC1 are displayed in green and marked with dashed arrows, while those with a negative contribution are displayed in red and marked with solid. Cross peaks in the loading plot are numbered as follows: 1, lupulone; 2, humulone; 3, colupulone; 4, cohumulone; 5, humulone 4a 3 a 6 5 4 1 6 50 3 2 5 1 5 7 5 7 6 5 5 150 7 6 5 5 7 5 140 7 100 80 100ppm F1 (13C) 2 2 40 5 60 7 20 0 series; 6, lupulone series; 7, total humulones and lupulones. 5 180 7 7 3.5 3.0 220 200 3 4 2.5 F2 (1H) 2.0ppm 1.5 1.0 0.5 Supplementary Figure 7 Loading plot for PC2 components displaying the region from 1H -0.8 - 4.9 ppm and 13C -10 - 240 ppm showing cross peaks contributing to PC2 variance. Signals with positive contribution to PC2 are displayed as green dots and with solid arrows, while those with negative contribution are shown as red dots and with dashed arrows. Some correlation peaks are labelled and assigned to specific hop constituents as follows, (a) cross peaks for 2-methylbut-3-en-2-ol; (b) linalool; (c) fatty acids and (d) total bitter acids. Supplementary Figure 8 PCA scoring plots of PC1 and PC2 of hop resin samples using 2D-1H- HMBC versus 1D-1HNMR as datasets. Note the comparable positioning of hop samples placed in circles across PC1, especially for most distant samples with negative score values i.e. HHE & HSE; with positive score values i.e. HTU and HHS. TPE HPE ○ 20 HSE HTU 0 HHT EHM ENB CZAG -20 HHE HHS ▪ ATPE ATHM HHM ∆ -40 PC2 (18.1%) 40 2D-1H - HMBC -100 -50 PC1 (67.5%) 0 50 10 HHT CZAG EHM HSE ENB HTU HHS ▪ TPE HPE ○ ATPE 0 -10 PC2 (26.0%) 20 1D-1H-NMR HHM ∆ ATHM HHE -20 0 PC1 (61.3%) 20 40 Supplementary Figure 9 Loading plot showing cross peaks contributing to PC1 variance and quantification of H3-16/21 crosspeak assigned for total lupulones (L) across samples. a) Loading plot for PC1 components of the region from 1H 0.4-2.6 ppm and 13C 0-230 ppm showing cross peaks contributing to PC1 variance. Signals with positive contribution to PC1 are displayed in green, while those with a negative contribution are displayed in red. b) Bar plot of total lupulones (L) crosspeak intensities assigned for H3-16/21 appearing at 1H (1.56 ppm) and 13C (25.5 ppm). The HHE cultivar showed the highest level as denoted in red colour, versus the HTU cultivar exhibiting the lowest amounts. This is in full agreement with absolute quantifications of total lupulones using 1H-NMR, (Farag et al., 2012). a b high in HHE high in HTU 0 Most important loadings PC1 most importend loadings PC1 20 H1.58/C25.5 H1.56/C25.5 40 20 60 100 100 F1 Chemical Shift (ppm) 150 140 140 100 120 180 50 180 160 200 c) overlay spektrum and loadings TPE HTU HSE_D HSE_C HPE HSE_B HHT HHS HHM ENB HHE CZAG_B 1.0 CZAG_C 1.5 F2 (1H) ATPE 2.0 1.0 CZAG_A 2.5 1.5 F2 Chemical Shif t (ppm) ATHM 2.0 EHM 220 2.5 0 200 220 F1 (13C) 80 100 80 H1.56/C25.5 H1.56/C25.5 150 60 50 40