ddi12329-sup-0001
advertisement
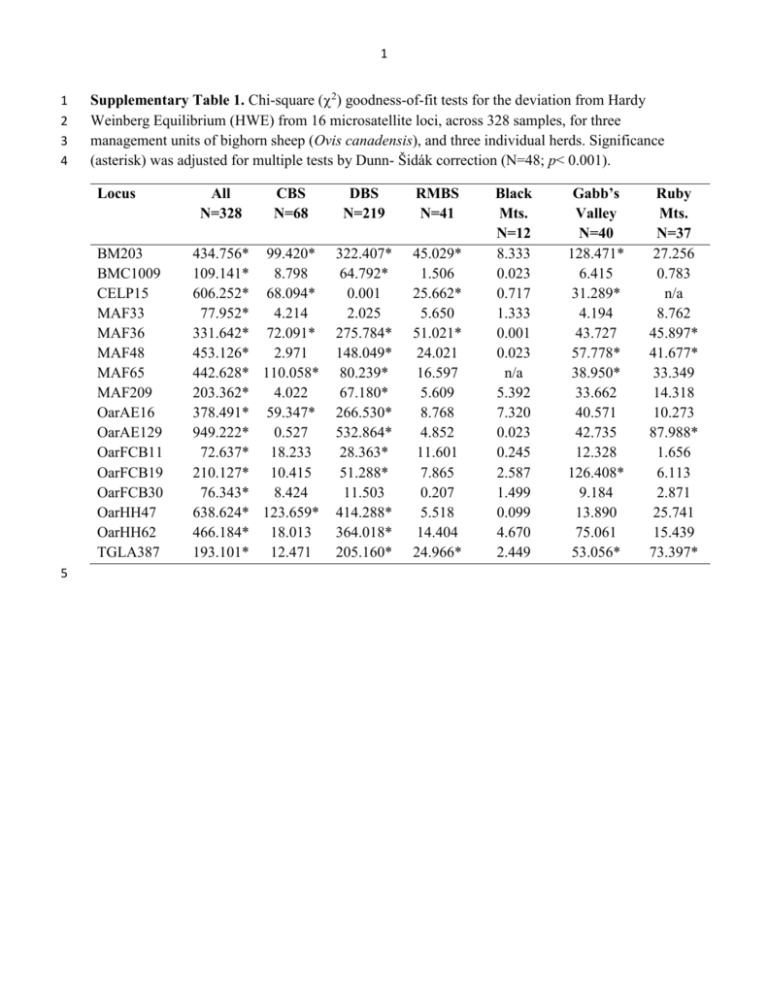
1 1 2 3 4 Supplementary Table 1. Chi-square (2) goodness-of-fit tests for the deviation from Hardy Weinberg Equilibrium (HWE) from 16 microsatellite loci, across 328 samples, for three management units of bighorn sheep (Ovis canadensis), and three individual herds. Significance (asterisk) was adjusted for multiple tests by Dunn- Šidák correction (N=48; p< 0.001). Locus BM203 BMC1009 CELP15 MAF33 MAF36 MAF48 MAF65 MAF209 OarAE16 OarAE129 OarFCB11 OarFCB19 OarFCB30 OarHH47 OarHH62 TGLA387 5 All N=328 CBS N=68 434.756* 99.420* 109.141* 8.798 606.252* 68.094* 77.952* 4.214 331.642* 72.091* 453.126* 2.971 442.628* 110.058* 203.362* 4.022 378.491* 59.347* 949.222* 0.527 72.637* 18.233 210.127* 10.415 76.343* 8.424 638.624* 123.659* 466.184* 18.013 193.101* 12.471 DBS N=219 RMBS N=41 322.407* 64.792* 0.001 2.025 275.784* 148.049* 80.239* 67.180* 266.530* 532.864* 28.363* 51.288* 11.503 414.288* 364.018* 205.160* 45.029* 1.506 25.662* 5.650 51.021* 24.021 16.597 5.609 8.768 4.852 11.601 7.865 0.207 5.518 14.404 24.966* Black Mts. N=12 8.333 0.023 0.717 1.333 0.001 0.023 n/a 5.392 7.320 0.023 0.245 2.587 1.499 0.099 4.670 2.449 Gabb’s Valley N=40 128.471* 6.415 31.289* 4.194 43.727 57.778* 38.950* 33.662 40.571 42.735 12.328 126.408* 9.184 13.890 75.061 53.056* Ruby Mts. N=37 27.256 0.783 n/a 8.762 45.897* 41.677* 33.349 14.318 10.273 87.988* 1.656 6.113 2.871 25.741 15.439 73.397* 2 6 7 8 9 10 11 12 Supplementary Table 2. Management Unit PCA (see Fig 5A) loadings from 600 (200 per unit CBS, DBS, RMBS) pseudo-presence samples of bighorn sheep (Ovis canadensis) within NBSMAs (see Fig. 1 and Fig 5B). Initially 26 variables were explored, but 13 variables were removed for strong correlation coefficients (r=|0.70|). The remaining variables were used in PCA and the first four components passed the Kaiser threshold criterion and together account for >75% of variation. Below the biplot (Gabriel, 1971) and loadings following Varimax rotation that were used for interpretation. 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 Variable bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr Comp.1 0.38449715 0.05182381 -0.03322271 0.22542144 0.20890510 -0.39915258 0.26659423 -0.40686580 -0.42796637 0.11444747 -0.11342299 -0.32984479 -0.20016364 Comp.2 0.12193380 -0.60448566 -0.53576179 -0.13837081 0.28393261 0.20290719 0.38811605 0.12514705 -0.04351716 0.01624897 -0.03414622 0.12815501 0.07556262 Comp.3 0.24729678 -0.13035163 -0.38436326 0.58368888 -0.41067820 -0.14732015 -0.28726426 0.07049304 -0.03957669 -0.18137175 0.26570767 0.07237932 0.21277626 Comp.4 -0.021516417 0.088448877 0.042942506 -0.053789086 0.004594864 -0.009639216 0.144354652 -0.078019959 -0.056831296 0.685883630 0.631123303 0.100616190 0.278981573 Comp.5 0.15621459 0.11408028 0.22529747 -0.15720608 0.29469143 -0.09363809 0.12289414 -0.11158650 -0.12360343 -0.56196760 0.22738387 0.12742867 0.60408399 Eigen Value 5.09977948 SD 2.2582691 Prop. Var. 0.3922907 2.19092858 1.4801786 0.1685330 1.43459346 1.1977452 0.1103533 1.04636733 1.02292098 0.08048979 0.91497630 0.95654394 0.07038279 Loadings: bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr Comp.1 Comp.2 Comp.3 Comp.4 0.400 0.250 0.130 -0.588 0.173 -0.658 0.362 0.202 0.489 -0.525 -0.108 -0.450 -0.117 0.138 0.170 -0.524 -0.401 0.136 0.112 -0.416 0.111 0.114 -0.174 -0.325 0.606 0.125 0.683 -0.316 0.117 0.156 -0.140 0.125 0.123 0.344 3 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 Supplementary Table 3. Nevada Bighorn Sheep Management Areas (NBHSMA, N=600) and random background samples (N=600) PCA results for 13 niche variables (see Supplementary Figure 2). Four components were sufficient to account for >70% of the variation. Loadings were those Eigenvectors that were most important after Varimax rotation, depicted in the biplot (below), and used for interpretation. bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr PC1 -0.44184262 -0.02801914 0.14778156 -0.30415596 -0.19654533 0.31636551 -0.12882823 0.39538181 0.46152278 -0.13177481 0.01912755 0.30463554 0.23337214 Eigen Value 4.10703445 SD 2.0265820 Prop. Var. 0.3159257 PC2 0.132027469 -0.424185785 0.001567514 -0.108434130 0.287663410 0.388295582 0.567595688 -0.114387087 -0.104504400 -0.334920448 0.266160559 0.139152274 0.093830207 PC3 -0.09268205 0.49889469 0.69121131 -0.26610613 0.25037132 0.05992710 0.13618124 -0.26857204 -0.05623956 -0.09534908 -0.11204582 -0.06457505 -0.08942433 PC4 0.049972670 -0.188391258 -0.156106572 -0.294595569 0.519649186 -0.171646467 -0.068762601 0.043373856 0.001480063 0.082139578 -0.647185216 0.081951526 0.335312428 PC5 -0.071809722 0.016406455 -0.026867232 -0.300122144 0.118915457 -0.114084055 0.055778084 -0.185382899 0.004404174 0.698142984 0.499081244 0.101271960 0.303947118 2.32599031 1.5251198 0.1789223 1.59344509 1.2623173 0.1225727 1.11876107 1.05771502 0.08605854 0.88892460 0.94282798 0.06837882 Loadings: bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr PC1 PC2 -0.435 -0.240 0.219 -0.209 -0.149 -0.309 0.113 0.198 0.494 -0.284 0.524 0.455 0.475 -0.379 0.402 0.268 0.171 0.213 PC3 PC4 -0.184 0.631 0.683 -0.161 -0.412 0.588 -0.179 -0.583 0.116 -0.175 0.326 4 58 59 60 61 Supplementary Table 4. Translocated bighorn sheep (Ovis canadensis) and source herds (British Columbia and Colorado) PCA eigenvectors for 13 niche variables (see Fig. 6). Two components were sufficient to account for >80% variation. Below are the biplot and loadings following Varimax rotation and used for interpretation. 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 Variable bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr Comp.1 0.3371871 -0.3334024 0.3206005 0.3190291 -0.1302214 -0.3140763 0.2130467 0.1852279 -0.3081962 0.3251032 -0.1387812 0.3242564 -0.2240631 Comp.2 -0.04903508 -0.10683417 -0.16466409 -0.07449225 -0.59437432 0.20217148 0.17301062 0.54270180 -0.01425693 0.03435445 0.46936764 -0.07818380 0.08428769 Comp.3 -0.02261678 -0.10561384 0.20988483 0.26423151 -0.11799994 -0.04216249 -0.72902767 -0.13394187 -0.22107642 -0.25408390 0.42388142 0.06506532 -0.10953754 Comp.4 -0.13934742 0.11557372 -0.15780334 -0.01332354 0.03857559 -0.07964365 0.27929035 -0.14327333 -0.16788623 -0.12028639 0.21065505 -0.25139149 -0.82798323 Comp.5 0.041418657 0.019892601 0.035869101 -0.034535808 0.257452520 -0.286612557 0.372496005 -0.322125487 -0.238136923 0.065447345 0.614293986 -0.006500954 0.407517965 Eigen Value 8.393889133 2.015665682 0.814878555 0.669203091 0.443194740 SD 2.8972209 1.4197414 0.90270624 0.81804834 0.6657287 Prop. Var. 0.6456838 0.1550512 0.06268297 0.05147716 0.0340919 Loadings: bio1 bio2 bio3 bio8 bio9 bio12 bio15 bio18 elev LC14 LC24 ndvi_sd_ca slope_degr PC1 PC2 PC3 0.293 0.176 -0.310 -0.179 0.409 0.411 -0.619 -0.338 0.135 -0.181 0.755 0.532 0.252 -0.361 -0.106 0.160 0.380 0.510 -0.397 0.328 -0.262 5 81 82 Supplementary Table 5. Multiplex PCR design with fluorescently labeled microsatellite primers used for genotyping bighorn sheep (N=328) in Nevada with locus-specific number of observed alleles (k) and range, plus observed (Ho) and expected (He) heterozygosity. Multiplex - Ta A - 59° B - 48° C - 59° D - 63° 83 84 Locus Label k Range Ho He OarFCB304 VIC 7 128-142 0.588 0.697 OarFCB193 PET 8 105-121 0.652 0.760 OarHH62 6-FAM 15 98-128 0.784 0.854 MAF33 NED 6 117-131 0.609 0.677 BMC1009 PET 7 278-290 0.364 0.430 TGLA387 PET 7 133-145 0.563 0.796 MAF209 NED 7 107-121 0.511 0.686 OarFCB266 6-FAM N/A OarCP20 VIC N/A MAF36 6-FAM 9 86-102 0.591 0.804 OarHH47 VIC 9 120-140 0.396 0.766 MAF48 PET 9 120-136 0.488 0.641 CELJP15 NED 5 159-169 0.123 0.307 OarAE129 6-FAM 15 163-191 0.543 0.852 MAF65 VIC 11 110-136 0.558 0.758 OarFCB11 NED 5 119-127 0.409 0.616 BM203 6-FAM 16 215-249 0.676 0.886 OarAE16 PET 11 84-104 0.591 0.800 Forward/Reverse 5’-3’ CCCTAGGAGCTTTCAATAAAGAATCGG CGCTGCTGTCAACTGGGTCAGGG TTCATCTCAGACTGGGATTCAGAAAGGC GCTTGGAAATAACCCTCCTGCATCCC TAATGAGTCAAACACTACTGAGAGAC AATATATAAAGAGAAAAGCTGGGGTGCC GATCTTTGTTTCAATCTATTCCAATTTC GATCATCTGAGTGTGAGTATATACAG GCACCAGCAGAGAGGACATT ACCGGCTATTGTCCATCTTG CAAAGTCTTAGAATAAACTGGATGG GTCCCTTTGTTTACTTTGATAAAAC TCATGCACTTAAGTATGTAGGATGCTG GATCACAAAAAGTTGGATACAACCGTGG GGCTTTTCCACTACGAAATGTATCCTCAC GCTTGGAAATAACCCTCCTGCATCCC GATCCCCTGGAGGAGGAAACGG GGCATTTCATGGCTTTAGCAGG CATATACCTGGGAGGAATGCATTACG TTGCAAAAGTTGGACACAATTGAGC TTTATTGACAAACTCTCTTCCTAACTCCACC GTAGTTATTTAAAAAAATATCATACCTCTTAAGG GGAAACCAAAGCCACTTTTCAGATGC AGACGTGACTGAGCAACTAAGTACG GGAAATACCTTATCTTTCATTCTTGACTGTGG CCTTCTTTCTCATTGCTAACTTATATTAAATATCC AATCCAGTGTGTGAAAGACTAATCCAG GTAGATCAAGATATAGAATATTTTTCAACACC AAAGGCCAGAGTATGCAATTAGGAG CCACTCCTCCTGAGAATATAACATG GGCCTGAACTCACAAGTTGATATATCTATCAC GCAAGCAGGTTCTTTACCACTAGCACC GGGTGTGACATTTTGTTCCC CTGCTCGCCACTAGTCCTTC CTTTTTAATGGCTCGGTAATATTCCTC CATCAGAGGAATGGGTGAAGACGTGG Reference Buchanan & Crawford, 1993 Buchanan & Crawford, 1993 Ede et al., 1994 Buchanan & Crawford, 1992 Bishop et al., 1994 Georges & Massey, 1992 Buchanan & Crawford, 1992 Buchanan & Crawford, 1993 Ede et al., 1995 Swarbrick et al., 1991 Henry et al., 1993 Buchanan et al., 1991 unp. see Marshal et al., 1998 Penty et al., 1993 Buchanan et al., 1992 Buchanan & Crawford, 1993 Bishop et al., 1994 Penty et al., 1993 (Buchanan et al., 1991; Swarbrick et al., 1991; Buchanan & Crawford, 1992; Buchanan et al., 1992; Georges & Massey, 1992; Buchanan & Crawford, 1993; Henry et al., 1993; Penty et al., 1993; Bishop et al., 1994; Ede et al., 1994; Marshall et al., 1998) 6 Supplementary Figure 1. Comparison of PCoA scatter plots of putative identifications (based on management units) and STRUCTURE assignments for K=2 and K=3 of bighorn sheep (Ovis canadensis) in the Great Basin and northern Mojave Deserts. In the lower two panels (K=2, K=3), squares show individuals that failed the assignment test for potentially admixed (open grey) and high statistical confidence hybrids (black). Blue=CBS, Green=DBS, Orange=RMBS. 7 Supplementary Figure 2. Test for spatial autocorrelation. PCA of Nevada Bighorn Sheep Management Areas (NBSMAs; see Fig. 1, N=600, black symbols) versus randomized background samples (N=600, grey symbols). Two axes account for 50% of the variation. Strong loadings on PC1 included mean annual temperature (+) and elevation (-), and PC2 were seasonality of precipitation (+) and mean diurnal temperature range (-). 8 SUPPLEMENTRY METHODS Sampling Tissues Fresh tissues were the primary source of genetic material and consisted of samples obtained from hunter-harvest return (muscle) and the NDOW disease-monitoring program (blood). We obtained samples from 55 herds representing the three management units maintained in Nevada. We use the term “management units” to refer to traditionally recognized subspecies (i.e., CBS, DBS, and RMBS) that are commonly used by natural-resource agencies. To augment some poorly sampled locations, we acquired fresh fecal pellets from individuallyobserved bighorn sheep. We extracted genomic DNA using DNeasy® Blood & Tissue Kit and modified protocols (Wehausen et al., 2004) for QIAamp® Stool Kit (Qiagen Valencia, CA, USA). Genetic data We generated multi-locus genotypes for 18 microsatellite loci previously used in bighorn sheep using multiplex (QIAGEN MULTIPLEX PCR KIT, Qiagen) polymerase chain reactions (PCR) with fluorescently labeled primers (Supplementary Table 5). We performed a minimum of two replicate PCRs per sample and per locus to detect genotyping errors that may result from degraded DNA (Taberlet et al., 1999). Consensus genotypes were those observed in both runs, but if we observed an allele only once, we conducted an additional set of PCRs. In all reactions, we included two negative controls, which did not include template DNA, to test for contamination of PCR reagents, and six positive control samples (known genotypes) to standardize analyses. We combined fluorescently labeled amplicons with ABI GENESCANTM 500 LIZ allelic ladder (APPLIED BIOSYSTEMS, Foster City, California) and analyzed them on an ABI 3730 DNA Analyzer at the Nevada Genomics Center (NGC). We scored individual peaks with GENEMARKER® v.1.85 (SOFTGENETICS LLC; State College, Pennsylvania) software and verified all genotypes manually. Two loci failed to consistently amplify (OarFCB266 and OarCP20) so were removed from further analyses resulting in 16 loci for characterizing genetic variation. To provide an independent sequence-based perspective of differentiation among genetic units, we sequenced a segment (1181bp) of the mitochondrially encoded NADH dehydrogenase 5 (ND5) gene for 110 samples representing the geographic breadth and taxonomic units of sheep in the study area. The ND5 gene was amplified by double-stranded PCR using two novel primers designed for this study: ND5-L2 (5°-AGTAGTTATCCATTCGGTCTTAGG-3°) and ND5-R3A (5°-GGTCTTTGGAGTAGAATCCGGTGAG-3°). PCRs consisted of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30s at 94 °C (denaturation), 30s at 50 °C (annealing), and 60s at 72 °C (extension) concluding with a final extension of 5 min at 72 °C. PCRs consisted of 30 L total volume, containing 3.0 L of MgCl2-free buffer, 3.0 L of MgCl2 solution, 3.0 L of dNTPs (2.0 m), 0.6 L of each primer (10 m), 0.2 L Taq polymerase, 1.0 L of template (c. 50-100 ng double-stranded DNA), and 18.6 L of sterile water. Negative controls, were used 9 as checks for contamination of PCR reagents. We then purified PCR products using a 20% dilute aliquot of EXOSAP-IT (Affymetrix, Santa Clara California) and sequenced products in both directions using PCR primers plus an internal primer designed specifically for this study: ND5F2 (5°-CCAACACAGCAGCCTTACAAGC-3°). Sequencing reactions were conducted with ABI BIGDYE TERMINATOR CYCLE SEQUENCING KIT 3.1, and we assessed purified products on an ABI 3730 DNA Analyzer in the NGC. We used GENEIOUS v.6.1.4 (Biomatters, San Francisco, California) to inspect products and aligned sequences using MUSCLE (Edgar, 2004). To verify authentic mtDNA, we translated the protein-coding regions of nucleotides into amino acid sequences, and deposited sequences in GenBank. Genetic diversity We assessed the microsatellite dataset quality using the program MICRO-CHECKER (Van Oosterhout et al., 2004), which can detect genotyping errors, allelic dropout, and null alleles. We estimated indices of genetic diversity including mean number of alleles per locus (NA) and mean observed (HO), expected (HE), and unbiased (uHE) heterozygosity using GENALEX v.6.5 (Peakall & Smouse, 2006, 2012). We tested for Hardy-Weinberg equilibrium (HWE) deviation using the chi-squared (2) goodness-of-fit test within the genetic units (identified by Bayesian clustering analyses, see below), and at a fine-scale for three well-sampled herds (Black Rock N=12, Muddy N=45, and Ruby N=36). The nominal level of statistical significance (=0.05) was adjusted to 0.001068 given the large number of comparisons (N=48) using the Dunn-Šidák correction factor (1-(1-)1/n) (Dunn, 1961; Šidák, 1967) considered an appropriate assessment for genetic data (Weir, 1996). Niche data To assess niche variation among translocated bighorn sheep, we used a novel approach considering there are few historical locations (Hall, 1946; Hall, 1981; Shackleton, 1985) with high quality information suitable for characterizing the scenopoetic niche (Peterson et al., 2011). Our goals were to 1) characterize ecotypic differences among management units, 2) place those differences within the context of spatial autocorrelation, and 3) compare bioclimatic conditions of translocated areas to those of source ranges (British Columbia and Colorado). Consequently, we characterized the distribution of bighorn sheep in Nevada using a pseudo-presence technique. Pseudo-presence identifies points randomly from within the range of the focal taxon with random points generated using Geographical Information Systems (GIS). Datasets from pseudopresence records can be prone to over-prediction because of broad extent-of-occurrence, where samples may represent both suitable and unsuitable areas, thereby exaggerating the occupied conditions (Graham & Hijmans, 2006; Hurlbert & Jetz, 2007; Jetz et al., 2008). The concern of exaggerated distributions may be less of an issue with large-bodied and mobile mammals (Richmond et al., 2010). Because precise, individual location data are not available from throughout our study region, our analysis is based on the areas within the state of Nevada 10 allocated to bighorn sheep management (Nevada Bighorn Sheep Management Areas; NBSMAs). As such, our analysis does not reflect actual usage of these areas, but rather what is potentially available to translocated animals. We generated three sets of points using ARCGIS 10 (ESRI, 2011). First, we generated randomized localities from within the NBSMAs (obtained June 2013 from Western Association of Fish and Wildlife Agencies Wild Sheep Working Group - WAFWAWSWG; Fig. 1) with 5 km spacing and 200 samples per management unit (CBS, DBS, RMBS; N=600). We also generated 5000 random points across the study area (Nevada) to assess correlation among bioclimatic variables and identify spatial autocorrelation (McCormack et al., 2010), and then down-sampled to 600 points to equate sample variance, and then compared environmental variation between NBSMAs versus background. Finally, to compare characteristics of the NBSMAs with those available in source ranges, we generated 200 random points from source management areas in Colorado and British Columbia (obtained from WAFWAWSWG). Temperature, precipitation, elevation, and slope are considered important for the persistence of bighorn sheep (Epps et al., 2004; Epps et al., 2007) and likely important in Grinnellian niche space (Soberón, 2007) and consequently the scenopoetic niche (Peterson et al., 2011). Niche variables were extracted for all randomly generated points. Niche-based variables included 19 bioclimatic and elevation (Hijmans et al., 2005) from the WorldClim database (2.5 minute, http//www.worldclim.org). We used a 10 m digital elevation model (DEM) from ESRI to characterize the degree slope using spline interpolation. Because species likely respond to a complex set of environmental (abiotic) conditions and biotic associations (McCormack et al., 2010; Moritz & Agudo, 2013) we incorporated five biotic vegetation-based variables including two land cover classifications, percent tree-cover, plus Normalized Difference Vegetation Index (NDVI) and NDVI standard deviation (www.landcover.org). NDVI and NDVI-SD that we standardized across 2010-2012, and corresponded to the period of genetic sampling. Literature Cited Bishop, M.D., Kappes, S.M., Keele, J.W., Stone, R.T., Sunden, S., Hawkins, G.A., Toldo, S.S., Fries, R., Grosz, M.D. & Yoo, J. (1994) A genetic linkage map for cattle. Genetics, 136, 619-639. Buchanan, F. & Crawford, A. (1992) Ovine dinucleotide repeat polymorphism at the MAF33 locus. Animal genetics, 23, 186-186. Buchanan, F. & Crawford, A. (1993) Ovine microsatellites at the OARFCB11, OARFCB128, OARFCB193, OARFCB266 and OARFCB304 loci. Animal Genetics, 24, 145-145. Buchanan, F., Swarbrick, P. & Crawford, A. (1991) Ovine dinucleotide repeat polymorphism at the MAF48 locus. Animal genetics, 22, 379-380. Buchanan, F., Swarbrick, P. & Crawford, A. (1992) Ovine dinucleotide repeat polymorphism at the MAF65 locus. Animal genetics, 23, 85-85. Dunn, O.J. (1961) Multiple comparisons among means. Journal of American Statistical Association, 56, 52-64. Ede, A., Peirson, C., Henry, H. & Crawford, A. (1994) Ovine microsatellites at the OarAE64, OarHH22, OarHH56, OarHH62 and OarVH4 loci. Animal genetics, 25, 51-51. 11 Edgar, R.C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. Bmc Bioinformatics, 5, 1-19. Epps, C.W., McCullough, D.R., Wehausen, J.D., Bleich, V.C. & Rechel, J.L. (2004) Effects of climate change on population persistence of desert-dwelling mountain sheep in California. Conservation Biology, 18, 102-113. Epps, C.W., Wehausen, J.D., Bleich, V.C., Torres, S.G. & Brashares, J.S. (2007) Optimizing dispersal and corridor models using landscape genetics. Journal of Applied Ecology, 44, 714-724. ESRI (2011) ArcGIS Desktop: Release 10. Environmental Systems Research Institute. Gabriel, K.R. (1971) The biplot graphic display of matrices with application to principal component analysis. Biometrika, 58, 453-467. Georges, M. & Massey, J.M. (1992) Polymorphic DNA markers in Bovidae. World Intellectual Property Organization. Graham, C.H. & Hijmans, R.J. (2006) A comparision of methods for mapping species ranges and species richness. Global Ecology and Biogeography, 15, 578-587. Hall, E.R. (1946) Mammals of Nevada. University of California Press, Berkeley and Los Angeles, California. Hall, E.R. (1981) The mammals of North America, 2nd edn. John Wiley & Sons, New York, New York. Henry, H., Penty, J., Pierson, C. & Crawford, A. (1993) Ovine microsatellites at the OARHH35, OARHH41, OARHH44, OARHH47 and OARHH64 loci. Animal genetics, 24, 222-222. Hijmans, R.J., Cameron, S.E., Parra, J.L., Jones, P.G. & Jarvis, A. (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978. Hurlbert, A.H. & Jetz, W. (2007) Species richness, hotspots, and the scale dependence of range maps in ecology and conservation. Proceedings of the National Academy of Sciences of the United States of America, 104, 13384-13389. Jetz, W., Sekercioglu, C.H. & Watson, J.E.M. (2008) Ecological correlates and conservation implications of overestimating species geographic ranges. Conservation Biology, 22, 110-119. Marshall, T., Slate, J., Kruuk, L. & Pemberton, J. (1998) Statistical confidence for likelihood‐based paternity inference in natural populations. Molecular ecology, 7, 639-655. McCormack, J.E., Zellmer, A.J. & Knowles, L.L. (2010) Does niche divergence accompany allopatric divergence in Aphelocoma jays as predicted under ecological speciation?: Insights from tests with niche models. Evolution, 64, 1231-1244. Moritz, C. & Agudo, R. (2013) The future of species under climate change: resilience or decline? Science, 341, 504-508. Peakall, R. & Smouse, P.E. (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. Peakall, R. & Smouse, P.E. (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics, 28, 2537-2539. Penty, J., Henry, H., Ede, A. & Crawford, A. (1993) Ovine microsatellites at the OarAEl6, OarAE54, OarAE57, OarAE119 and OarAE129 loci. Animal genetics, 24, 219-219. Peterson, A.T., Soberón, J., Pearson, R.G., Anderson, R.P., Martínez-Meyer, E., Nakamura, M. & Araújo, M.B. (2011) Ecological niches and geographic distributions (MPB-49). Princeton University Press. Richmond, O.M.W., McEntee, J.P., Hijmans, R.J. & Brashares, J.S. (2010) Is the climate right for pleistocene rewilding? Using species distribution models to extrapolate climatic suitability for mammals across continents. Plos ONE, 5, e12899. Shackleton, D.M. (1985) Ovis canadensis. Mammalian Species, 230, 1-9. Šidák, Z. (1967) Rectangular confidence regions for the means of multivariate normal distributions. Journal of American Statistical Association, 62, 626-633. 12 Soberón, J. (2007) Grinnellian and Eltonian niches and geographic distributions of species. Ecology Letters, 10, 1115-1123. Swarbrick, P., Buchanan, F. & Crawford, A. (1991) Ovine dinucleotide repeat polymorphism at the MAF36 locus. Animal genetics, 22, 377-378. Taberlet, P., Waits, L.P. & Luikart, G. (1999) Noninvasive genetic sampling: look before you leap. Trends in Ecology & Evolution, 14, 323-327. Van Oosterhout, C., Hutchinson, W.F., Wills, D.P.M. & Shipley, P. (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535-538. Wehausen, J.D., Ramey, R.R.I. & Epps, C.W. (2004) Experiments in DNA extraction and PCR amplificaiton from bighorn sheep feces: the importance of DNA extraction method. Journal of Heredity, 95, 503-509. Weir, B.S. (1996) Genetic Data Analysis II: Methods for discrete population genetic data. Sinauer Associates, Inc., Sunderland, Massachusetes.