Nanobots_revised_proposal-1
advertisement

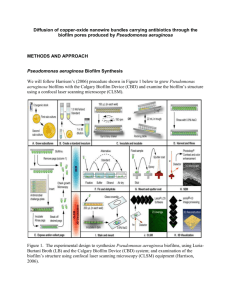
Diffusion of copper-oxide nanowire bundles carrying antibiotics through the biofilm pores produced by Pseudomonas aeruginosa INTRODUCTION AND BACKGROUND We propose to study the control of Pseudomonas aeruginosa infections by using nanowires to deliver antibiotics across the Pseudomonas aeruginosa’s biofilm to treat chronic infections. According to previous research, among 40 patients suffering from cystic fibrosis, 77% were infected by Pseudomonas aeruginosa, even though 71% of these patients exhibited a normal immune response (Neter 1974; Diaz et al. 1970; Diaz and Neter 1970). Consequently, conventional antibiotic therapy was ineffective against the Pseudomonas aeruginosa infection. Thus, we will use copper-oxide nanowire bundles (CuO NWB) to penetrate the biofilm produced by Pseudomonas aeruginosa to deliver antibiotics and kill the bacteria residing inside the biofilm. Cystic fibrosis is an inherited disease that causes a thick, sticky mucus build up in the lungs, which is an ideal environment for Pseudomonas aeruginosa biofilms to fluorish. Pseudomonas aeruginosa infections form biofilms to defend themselves against human immunity and conventional antibiotic therapy (Costerton et al. 1999; Nickel et al. 1985). This is important because a Pseudomonas aeruginosa’s biofilm forms a 200 µm barrier of sugars and carbohydrates to prevent antibiotics from reaching the pathogens (Hanlon et al. 2001; Anwar and Costerton 1992; Costerton et al. 1987; Govan and Deretic 1996). Earlier studies have used diffusion as a way to enter the pathogen’s biofilm layer to kill it, which is the method we propose to use in this study. This is because Pseudomonas aeruginosa’s biofilm consists of numerous pores and channels, which can be used for the diffusion of nanowires (Costerton et al. 1999; Stewart 1996). Nanowires have been used in various biological applications due to their size and selectivity (Li et al. 2010; Bao et al. 2008; Lu et al. 2007). These wires have a diameter of the nanometer scale, which can have different shapes and properties depending on the material they are made of. And, we can arrange these nanowires into nanowire bundles to form a synthetic mesh-like net that will increase the available surface area for antibiotic attachment. Metal ions have been used by Harrison (2005) to kill the pathogen by diffusing metal ions through the Pseudomonas aeruginos’s biofilm. In addition, the results showed that a high concentration and long exposure time of various metal ions were able to completely eliminate the biofilm (Harrison et al. 2005). And, the lengthy exposure time was a result of cationic binding of the metal ions to the biofilm that restricted the rate of penetration (Harrison et al. 2005). Harrison (2005) also determined that 40 mM of lead, 120 mM of zinc, 140 mM of cobalt and nickel, and 300 mM of aluminum metal ions were required to destroy a biofilm, whereas only 30 mM of copper was required to have the same outcome. Thus, we decided to make our nanowires out of copper because copper was effective at low concentrations and non toxic to biological systems. Therefore, we will use electrostatics to compose CuO NWB antibiotic carriers. We predict the antibiotics will occupy the positive charge on the CuO NWBs and will not adhere to the surface of the biofilm during diffusion. Previous research performed by Walters et al. (2002) demonstrated tobramycin and ciprofloxacin eventually penetrated the biofilm, but failed to kill Pseudomonas aeruginosa once inside the biofilm. As indicated by Figure 1, both ciprofloxacin and tobramycin did not increase Pseudomonas aeruginosa’s mortality rate over a 100 hour exposure time. Figure 1. Killing of P. aeruginosa in biofilms in exposure to ciprofloxacin (A) and killing of P. aeruginosa in biofilms in exposure to tobramycin (B). Filled squares were the treatment and the unfilled were the controls (Walter et al. 2002) Thus, the ineffectiveness of these antibiotics was not because of poor penetration, but oxygen limitation and low metabolic activity inside the biofilm (Walters et al. 2002). A lack of oxygen within the biofilm either prevented Pseudomonas aeruginosa growth or the action of the antibiotics; whereas, free swimming bacterial cells were susceptible to tobramycin and ciprofloxacin (Walters et al. 2002). For these reasons, we decided to use these two antibiotics in conjunction with our CuO NWBs to kill the pathogen inside the biofilm. These bundles will not block the Pseudomonas aeruginosa biofilm pores because of their nanometer size. Therefore, oxygen transport will be restored, metabolic activity will continue as normal, and we will be able to spread the antibiotics over a larger area. So, by combining these antibiotics to our CuO NWBs we are creating a synthetic neutrophil extracellular trap (NET). According to Brinkmann (2004), white blood cells in a normal immune response form a neutrophil net by secreting proteins and chromatin. This net traps the pathogen and prevents damage to the adjacent tissues at the site of infection. NETs bind to bacterial cells, prevent the spread of infection, and kill the pathogen using antimicrobial agents (toxins, chemicals, or antibodies) (Brinkmann et al. 2004; Weinrauch et al. 2002). As a result of these above 2. Neutrophil advantageous properties, we will use a bundle of nanowires to imitate a Figure NET and trap the extracellular trap (NET). pathogen. According to Tirouvanziam (2007), NETs are rapidly killed by toxins released by the bacterial cells making them ineffective. This is why patients with cystic fibrosis are continuously suffering from recurring episodes of Pseudomonas aeruginosa infection. So, could CuO NWBs carrying antibiotics diffuse through the porous structure of Pseudomonas aeruginosa’s biofilm and act as a synthetic NET? We hypothesize that CuO NWBs will imitate a synthetic NET and carry antibiotics through the biofilm of Pseudomonas aeruginosa to kill the pathogen from the inside. If this synthetic NET carrying antibiotics diffuses through the biofilm successfully and if the antibiotics detach from the NET inside the biofilm, we predict no growth on the antibiotic/CuO NWB agar plates. PROPOSED RESEARCH Experimental synopsis Pseudomonas aeruginosa Biofilm Synthesis We will follow Harrison’s (2006) procedure shown in Figure 1 to grow Pseudomonas aeruginosa biofilms using the Calgary Biofilm Device (CBD). We will use a confocal laser scanning microscope (CLSM) and its 3D imaging properties to examine the biofilm’s structure and confirm the presence of pores. Figure 1. The experimental design to synthesize Pseudomonas aeruginosa biofilms, using LuriaBurtani Broth (LB) and the Calgary Biofilm Device (CBD) system; and examination of the biofilm’s structure using confocal laser scanning microscopy (CLSM) equipment (Harrison, 2006). We will follow Harrison et al. 2006 procedure on the creation of Pseudomonas aeruginosa biofilm with the exception of using a scanning electron microscopy. Synthesis of copper oxide nanowires We will follow the methods outlined by Li et al. 2010 to synthesize the copper oxide nanowire bundles. We will create a one dimensional nanowire and combine it with an aqueous solution of CuCl2. This composite solution will be vacuum filtered and dried in an oven to produce our nanowires composition. The initial template will be removed with a solution of NaOH and the copper oxide nanowire bundles will dry a final time. Creating our copper nanowire carriers takes 24 hours to complete. We will get a visual representation of our carriers by using transmission electron microscopy (TEM) to generate an image. X-ray diffraction (XRD) analysis will be used to determine the structure and correct chemical composition of our carriers. We will look for successful assembly of the copper oxide nanowire bundles. Antibiotic Coupled Nanowire Synthesis We will use electrostatic interactions to carry our two types of antibiotics, Ciproflaxin and Tombramycin, with our copper oxide nanowire carrier. We will synthesize the nanowire carriers following the methods outlined in Li (2010), but we will reduce the copper oxide to form a charged Cu2+ nanowire carrier. Our antibiotics will carry an opposite charge by salvation and interact with the charged carriers by the method of electrostatics. We will use Fourier transformIR (FT-IR) to get spectra information on our copper-oxide nanowires, antibiotic, and the combination of nanowires and antibiotics. Controls and Test Samples Pseudomonas aeruginosa biofilms will be exposed to each parameter listed in Table 1 below: Table 1. Experimental group parameters tested with Pseudomonas aeruginosa biofilms. EXPERIMENTAL GROUP EXPERIMENTAL TREATMENT PARAMETERS EXPECTED RESULT A Control B Antibiotic only C Nanowire only D Nanowire + Antibiotic No antibiotic No copper-oxide nanowire Antibiotic No copper-oxide nanowire No antibiotic Copper-oxide nanowire Antibiotic fused Copper-oxide nanowire Growth Growth Limited growth ( copper-oxide toxicity) No growth (antibiotic penetration) To prepare each of our test group parameters in Table 1 above, we will follow the procedures outlined in Table 2 below: Table 2. Experimental treatment group preparations. TEST GROUP PREPARATION PROCEDURE Control Add 200 µL of 0.9% NaCl to 24 wells of a 96-well microtiter plate of a randomly selected region (1, 2, 3, or 4) (Fig. 2) Antibiotic only Add 200 µL of antibiotic to 24 wells of a 96-well microtiter plate of a randomly selected region (1, 2, 3, or 4) (Fig. 2) Nanowire only Add copper-oxide nanowires to 24 wells of a 96-well microtiter plate containing 200 µL of 0.9% NaCl of a randomly selected region (1, 2, 3, or 4) (Fig. 2) Nanowire + Antibiotic Add 200 µL of diluted antibiotic fused copper-oxide nanowires to 24 wells of a 96-well microtiter plate of randomly selected regions (A,B,C, and D) (Fig. 2) Figure 2. A microtiter plate illustrating the regions of the treatment parameters control (A), antibiotic only (B), nanowire only (C), and antibiotic fused nanowire (D) (1, 2, 3, and 4). Each treatment will consist of 24 wells out of the 96 (Ceri et al. 1999). Sample Size We expect little to no growth once we penetrate the biofilm layer of Pseudomonas aeruginosa with our synthetic neutrophil net. To determine the sample size (N) we will a chisquared logistical model to determine the noncentrality parameter (λ) using the formula N = λ / w2. The effect size (w) is the effect we are interesting in detecting. With our growth and no growth model we expect to see a medium effect size of 0.3. With 14 degrees of freedom in our experimental design, we obtained a noncentrality parameter of 27.2. The sample size that is required for our experiment is 303. We will need at least 13 MBEC plates to get the required sample size. Analysis and Interpretation We will use Harrison’s (2006) viable cell counting procedure to determine Pseudomonas aeruginosa growth following each treatment parameter discussed previously. To collect our results after conducting each treatment parameter, we will follow the procedure outlined in Figure 3 below. First, we will rinse the biofilms by placing the CBD peg lid into a 96-well microtiter plate containing 200 µL of 0.9% NaCl in each well for 2 min (Fig. 1D). Then, the CBD pegs will be removed using flamed pliers and placed in a microtiter plate containing 200 µL of 0.9% NaCl in each well (Fig. 1E). This will be followed by sonication: using an Aquasonic 250HT ultrasonic cleaner (60 Hz for 5 min.) to remove the bacterial cells from the peg surface. The bacterial cells will be serially diluted in 0.9% NaCl, plated on LB agar medium, and incubated at 35℃ for 24 hours. The following day, we will count the number of colony forming units (CFUs) on each plate and record the results. Figure 3. Diagram of experimental procedure used for group tests B, C, and D (Table 1). Step D above will incubate with antibiotics for group tests B and D and with copper oxide nanowires for group tests C and D (Herrmann, 2010). Conclusion Positive outcomes of results REFERENCES Anwar H, Costerton JW. 1992. Effective use of antibiotics in the treatment of biofilm-associated infections. American Society for Microbiology News. 58: 665–668. Bao SJ, Li CM, Zang JF, Cui XQ, Qiao Y, Guo J. 2008. New nanostructured TiO2 for direct electrochemistry and glucose sensor applications. Advanced Functional Materials. 18: 591-599. Brinkmann V, Reichard U, Goosmann C, Fauler B, UhlemannY, Weiss SD, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. American Association for the Advancement of Science. 303: 1532-1535. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. Journal of Clinical Microbiology. 37:1771-1776. Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annual Reviews of Microbiology. 41: 435–464. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: A common cause of persistent infections. Science. 284: 1318-22. Diaz F, Mosovich LL, Neter E. 1970. Serogroups of Pseudomonas aeruginosa and the immune response of patients with cystic fibrosis. Journal of Infectious Diseases. 121: 269-274. Diaz F, Neter E. 1970. Pseudomonas aeruginosa: Serogroups and antibody response in patients with neoplastic diseases. American Journal of the Medical Sciences. 259: 340-345. Govan JRW, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiological Reviews. 60: 539–574. Hanlon WG, Denyer Ps, Olliff JC, Ibrahim JL. 2001. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. American Society for Microbiology. 67: 2746-53. Harrison JJ, Turner RJ, Ceri H. 2005. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Biofilm Research Group. University of Calgary. 7: 981-94. Harrison JJ, Ceri H, Yerly J, Stremick CA, Hu Y, Martinuzzi R, Turner RJ. 2006. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biological Procedures Online. 8:194215. Herrmann G, Yang L, Wu H, Song Z, Wang H, Hoiby N, Ulrich M, Molin S, Riethmuller J, Doring G. 2010. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. The Journal of Infectious Diseases. 202:1585-1592. Li Y, Zhang Q, Li J. 2010. Direct electrochemistry of hemoglobin immobilized in CuO nanowire bundles. Talanta. 83: 162-66. Lu X, Zou G, Li J. 2007. Hemoglobin entrapped within a layered spongy Co3O4 based nanocomposite featuring direct electron transfer and peroxidase activity. Journal of Materials Chemistry. 17: 1427-1432. Neter E. 1974. Pseudomonas aeruginosa infection and humoral antibody response of patients with cystic fibrosis. The Journal of infectious diseases. 130: 132-133. Nickel JC, Ruseska I, Wright JB, Costerton JW. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Journal of Antimicrobial Agents and Chemotherapy. 27: 619-624. Stewart PS. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Journal of Antimicrobial Agents and Chemotherapy. 40: 2517-2522. Tirouvanziam.R, Gernez Y, Conrad KC, Moss BR, Schrijver I, Dunn EC, Davies AZ, Herzenberg AL, Herzenberg AL. 2007. Profound functional and signaling changes in viable inflammatory neutrophills. National Academy of Science. 105: 4335-4339. Walters CM, Roe F, Bugnicourt A, Franklin MJ, Stewart SP. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramyacin. American Society for Microbiology. 47: 317-23. Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. 2002. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 417: 91-94.