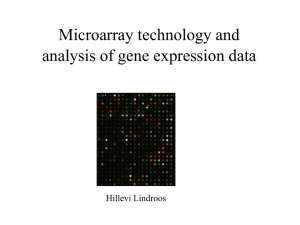
escalating-dose prednisolone treatment on an acute
advertisement

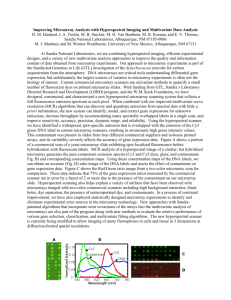
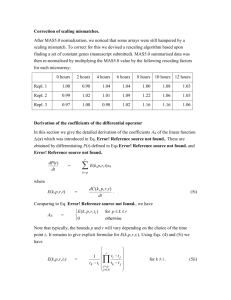
SUPPLEMENTARY DATA TO THE ARTICLE TITLED: “PREDNISOLONE EXERTS LATE MITOGENIC AND BIPHASIC EFFECTS ON RESISTANT ACUTE LYMPHOBLASTIC LEUKEMIA CELLS: RELATION TO EARLY GENE EXPRESSION” George I Lambrou1, Spiros Vlahopoulos1, Chrisanthi Papathanasiou1, Maria Papanikolaou1, Michael Karpusas2, Emmanouil Zoumakis3, Fotini Tzortzatou-Stathopoulou1 1 Hematology/Oncology Unit, First Department of Pediatrics, University of Athens Medical School, “Aghia Sophia” Children’s Hospital, Athens, Greece 2 University Research Institute for the Study and Treatment of Childhood Genetic and Malignant Diseases 3 Choremeio Research Laboratory, First Department of Pediatrics, University of Athens Medical School, “Aghia Sophia” Children’s Hospital, Athens, Greece MICROARRAY EXPERIMENTATION AND DATA ANALYSIS cDNA microarray chips (1,200 genes) were obtained from TAKARA (Human Cancer Chip v.40).[1, 2] Hybridization was performed with the CyScribe Post-Labeling kit (RPN5660, Amersham) as described by the manufacturer, utilizing the Cy3 and Cy5 fluorescent dyes. cDNAs were purified with QIAGEN PCR product clean-up kit (Cat # 28104). Slides were activated at 55o C for 30 min in 1% BSA. Samples were applied on the slides, and let to hybridize overnight at 55o C. The following day, slides were washed in 200 ml 0.1× SSC and 0,1% SDS for 3× 5 min, in 200 ml 0.1× SSC for 2× 5 min and in 200 ml ddH2O for 30 sec. Slides were dried by centrifugation at 1500 rpm for 3 min and scanned with a microarray scanner (ScanArray 4000XL). Images were generated with ScanArray microarray acquisition software (GSI Lumonics, USA). To obtain the highest possible experimental accuracy, cDNAs were used from three experimental setups, each one consisting of three independent experiments. In particular, the experimental setups consisted of the three following pairs: control (0 μM prednisolone) (Cy3) vs 10nM prednisolone (Cy5) (designated as 0vs1), 10nM prednisolone (Cy3) vs 701μM prednisolone (Cy5) (designated as 1vs3), control (0 μM prednisolone) (Cy3) vs 701μM prednisolone (Cy5) (designated as 0vs3). This is a ‘simple loop’ experimental design, taking into account all possible combinations between samples, as previously described.[3] Microarray Data Analysis and Statistics: Microarray data analysis was performed with ImaGene v.6.0 Software (BioDiscovery Inc, CA). Normalization was performed by dividing each data value with the median (50% percentile) and background correction was performed with global background correction, sub-grid based and negative control spots.[4-6] Genes were filtered first for their spot quality. This was done by acquiring a ‘0’ flag marking on each spot as assessed by ImaGene software (Signal-to-Noise Ratio, SNR) in at least two experiments for each concentration (notice that SNR is calculated by the software from the equation 1 SNR R ,G B , where μR,G and μB is the mean value intensity for the respective channel (Cy3 or Cy5) B and mean background intensity respectively and σB is the background mean signal standard deviation. A threshold of 2 has been set as a cut-off value, meaning that spot intensity for at least one channel should be twice as much as that of the background). The mean ratio of each triplicate of the ith gene was calculated from R ch2i 1 j log 2 ( ) , where R is the mean expression ratio of a gene among a triplicate, ch2i and j i 1 ch1i ch1i are the normalized intensities of the two channels (Cy5 and Cy3 respectively). Or equivalently stated it is the geometric mean where R log 2 (GM ) log 2 ( j j ch2i ch1 i 1 ), j 3 . Furthermore, genes with a i good spot quality were tested against their differential expression consistency between the experimental setups i.e. genes in the third experiment (“0vs3”) should manifest a behavior consistent to the previous two. For example, a gene that is 2-fold overexpressed in the “0vs1” experiment and 2-fold overexpressed in the “1vs3” experiment should be approximately 4-fold overexpressed in the “0vs3” experiment, in logarithmic scale. This was calculated by the observation that the experimental setup followed a pattern of additive expressions. More specifically, fold expression in the first two experiments should be reflected as a sum in the third experiment. To explain this in more detail an example is given: a two-fold expression in a gene between control and 10nM prednisolone (0vs1) should give a log2 of 1. An another two- or fourfold expression between 10nM prednisolone and 701μM prednisolone (1vs3) equals to log2 of 1 (21) and 2 (22) respectively. The fold expression for the comparison between control and 701μM prednisolone (0vs3) that should be observed will be then log2(22)=2 and log2(23)=3 respectively. This fold expression in the 0vs3 experimental setup equals to the sum of the previous two experiments. In other words it should be R0,3=R0,1+R1,3 where Ri,j (j=1,2,3) is the ratio of the ith gene in the jth experiment (practically the third vector R0,3 should be the sum of the vectors R0,1 and R1,3 ). Taking the standard deviation (σ) between the calculated R0,3 and the experimental R0,3 a criterion of the consistency between the experiments is drawn. Genes that did not conform to this rule, specifically those with σ>1, were excluded from further analysis. Furthermore, each gene was tested for its significance in differential expression using a z-test. Since there were 9 independent experiments (three for each concentration combinations i.e. 0vs1, 1vs3, 0vs3) a meta-analysis was performed where p-values were combined in order to obtain a single p-value. pvalue combination was performed using the Fisher’s transformation for combining tests of significance. pvalue combinations have a chi-square distribution which is given by the equation x22k 2 k ln( p ) with i 1 i 2k degrees of freedom. Genes were considered to be significantly differentially expressed if they obtained a p-value <0.01 in all experiments. The False Discovery Rate has been calculated as described previously.[7-9] There was a FDR of 0.5% for a combined p<0.01 for experimental setup 0vs1, 0.6% for a 2 combined p<0.01 for experimental setup 1vs3 and 0.9% for a combined p<0.01 for the experimental setup 0vs3. Clustering analysis and chromosome mapping [10, 11] were performed with Genesis 1.7.2 (Technische Universitaet-Graz, Austria)[12] using Pearson’s correlation (r) and Spearman rank order correlation (ρ). Pearson’s correlation was calculated by order correlation was calculated by and Spearman rank For clustering analysis hierarchical clustering by Euclidian distance was used.[13] Also, a methodology of Self Organizing Maps (SOMs) was used in order to determine patterns of expression and it was compared to the hierarchical clustering results. 3 REFERENCES [1] Kiguchi T, Niiya K, Shibakura M, et al. Induction of urokinase-type plasminogen activator by the anthracycline antibiotic in human RC-K8 lymphoma and H69 lung-carcinoma cells. International journal of cancer 2001; 93:792-797. [2] Miyazato A, Ueno S, Ohmine K, et al. Identification of myelodysplastic syndrome- specific genes by DNA microarray analysis with purified hematopoietic stem cell fraction. Blood 2001; 98:422-427. [3] Yang YH, Speed T. Design issues for cDNA microarray experiments. Nature reviews 2002; 3:579-588. [4] Zien A, Aigner T, Zimmer R, Lengauer T. Centralization: a new method for the normalization of gene expression data. Bioinformatics (Oxford, England) 2001; 17 Suppl 1:S323331. [5] Wu W, Xing EP, Myers C, Mian IS, Bissell MJ. Evaluation of normalization methods for cDNA microarray data by k-NN classification. BMC bioinformatics 2005; 6:191. [6] Kroll TC, Wolfl S. Ranking: a closer look on globalisation methods for normalisation of gene expression arrays. Nucleic acids research 2002; 30:e50. [7] Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical hypotheses 1995; 45:486-490. [8] Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America 2003; 100:9440-9445. [9] Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods in molecular biology (Clifton, NJ 2003; 224:149-157. 4 [10] Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole- genome expression data reveals chromosomal domains of gene expression. Nature genetics 2000; 26:183-186. [11] Reyal F, Stransky N, Bernard-Pierrot I, et al. Visualizing chromosomes as transcriptome correlation maps: evidence of chromosomal domains containing co-expressed genes--a study of 130 invasive ductal breast carcinomas. Cancer research 2005; 65:1376-1383. [12] Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics (Oxford, England) 2002; 18:207-208. [13] Quackenbush J. Computational analysis of microarray data. Nature reviews 2001; 2:418- 427. 5