spatial stats notes from Catherine R.
advertisement

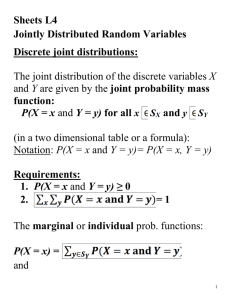
# Code from class—2.29.2010
library(nlme)
trees = read.csv(file.choose())
#The ‘treedbh.csv’ dataset
trees1 = na.exclude(trees)
#remove rows of missing values
trees2 = subset(trees1,trees1$dead==0) #ignore dead trees
trees3 = subset(trees2,duplicated(cbind(trees2$xcoord,trees2$ycoord))==F) #remove ‘double stems’ with same cords.
tr.mod1 = gls(INCR~DBH,data=trees3) # model of tree growth (INCR) as a function of stem size
plot(Variogram(tr.mod1,form=~xcoord+ycoord), ylim=c(.8,1.2))
#Original variogram using built in Variogram function in nlme
#######################################33
#Alternative using the geoR package
library(geoR)
#str(trees3)
trees4 = trees3
#create geoR object, includes x,y coords and response (in this case model residuals)
trees4$coords <- cbind(trees4$xcoord, trees4$ycoord)
trees4$data = tr.mod1$resid
#Plot empirical semivariograms
par(mfrow=c(1,2))
# all distances
plot(variog(trees4, option="cloud", max.dist=140))#each point corresponds to one pair
plot(variog(trees4), ylim=c(1,1.6))#averaged for each bin
tree.vario= variog(trees4)
#Standard error
yplus = tree.vario$v+1.96*sqrt(2) * tree.vario$v/sqrt(tree.vario$n)
yminus = tree.vario$v-1.96*sqrt(2) * tree.vario$v/sqrt(tree.vario$n)
library(Hmisc)
errbar(tree.vario$u,tree.vario$v, yplus, yminus, add=T)
#empirical semivariogram continued
#General rule ~1/2 max. distance
tree.vario= variog(trees4, max.dist=140)
#error
yplus = tree.vario$v+1.96*sqrt(2) * tree.vario$v/sqrt(tree.vario$n)
yminus = tree.vario$v-1.96*sqrt(2) * tree.vario$v/sqrt(tree.vario$n)
plot(tree.vario, ylim=c(1,1.6))
errbar(tree.vario$u,tree.vario$v, yplus, yminus, add=T)
################################
#Variogram model fitting by WLS
### geoR library function variofit: variogram models fitted by weighted or ordinary least squares.
# Use weights argument to define the loss function to be minimised.
# default: weights = “npairs”: weights are given by the number of pairs in each bin.
#Other arguments:
#weights= “cressie”: weights as suggested by Cressie (1985).
#LOSS(theta) = sum_k n_k [(hat(gamma_k) - gamma_k(theta))/{gamma_k(theta)}]^2
#weights="equal": equal values for the weights, corresponds to the ordinary least squares
tree.wls = variofit(tree.vario, ini.cov.par=c(1.35, 30), nugget=1, cov.model="exponential", weights="cressie")
summary(tree.wls)
#$spatial.component
#sigmasq (sill)
phi (RANGE)
#52.94009
19340.13725
#$spatial.component.extra
#kappa
# 0.5
#$nugget.component
# tausq
#1.204582
#$practicalRange
#[1] 57937.87
#$sum.of.squares
# value
#385.6431
########$estimated.pars
#
tausq (nugget)
# 1.204582
sigmasq (sill)
52.940087
phi (range)
19340.137246
# Create envelopes
wls.env = variog.model.env(trees4, obj.variog=tree.vario, model.pars=tree.wls)
par(mfrow=c(1,1))
plot(tree.vario, envelope=wls.env, ylim=c(1,1.6))
errbar(tree.vario$u,tree.vario$v, yplus, yminus, add=T)
lines(tree.wls, lty=2)
#You can specify different covariance models—spherical, exponential, gaussian, etc—see cov.spatial #documentation
# Example using the wave covariance model
tree.wls.wave <-variofit(tree.vario,c(1.35, 30),cov.model="wave",weights="cressie")
summary(tree.wls.wave)
#$pmethod
#[1] "WLS (weighted least squares)"
#$cov.model
#[1] "wave"
#$spatial.component
#sigmasq
phi
#0.2607983 34.7699407
#$spatial.component.extra
#kappa
#0.5
#$nugget.component
# tausq
#1.250420
#$fix.nugget
#[1] FALSE
#$fix.kappa
#[1] TRUE
#$practicalRange
#[1] 104.0128
#$#sum.of.squares
# value
#341.2557
#$estimated.pars
# tausq sigmasq
phi
# 1.2504204 0.2607983 34.7699407
# The wave model- envelopes
wls.env.wave = variog.model.env(trees4, obj.variog=tree.vario, model.pars=tree.wls.wave)
par(mfrow=c(1,1))
plot(tree.vario, envelope=wls.env.wave, ylim=c(1,1.6))
errbar(tree.vario$u,tree.vario$v, yplus, yminus, add=T)
lines(tree.wls.wave, lty=2)
#
tausq (nugget)
#WLS (exp) : # 1.204582
#WLS(wave)
# 1.2504204
sigmasq (sill)
phi (range)
52.940087
0.2607983
19340.137246
34.7699407
##############################################################
################################################################
#Likelihood based methods:
# library geoR function likfit: Maximum likelihood (ML) or restricted maximum likelihood (REML) parameter estimation
# Arguments
# lik.method = "ML" for maximum likelihood (default) or "REML" for restricted maximum likelihood.
#trend = “cte” (default), assumes the mean is constant over the region. See help(trend.spatial) for other options
# cov.model: specifies the model for the correlation function. Defaults are equivalent to the exponential model.
#Fit via Maximum likelihood
tree.ml = likfit(trees4, ini.cov.par=c(.35, 30), nugget=1, lik.method="ML")#
#Sensitive to starting parameters (sill, range), so need to run multiple times with different starting values
#Takes ~15minutes/run
summary(tree.ml)
#Summary of the parameter estimation
#----------------------------------#Estimation method: maximum likelihood
#Parameters of the mean component (trend):
# beta
#-0.078
#Parameters of the spatial component:
# correlation function: exponential
#
(estimated) variance parameter sigmasq (partial sill) = 0.2843
#
(estimated) cor. fct. parameter phi (range parameter) = 26.19
# anisotropy parameters:
# (fixed) anisotropy angle = 0 ( 0 degrees )
# (fixed) anisotropy ratio = 1
#Parameter of the error component:
#
(estimated) nugget = 1.142
#Transformation parameter:
# (fixed) Box-Cox parameter = 1 (no transformation)
#Practical Range with cor=0.05 for asymptotic range: 78.4482
#Maximised Likelihood:
# log.L n.params
AIC
BIC
#"-1734"
"4" "3477" "3497"
#non spatial model:
# log.L n.params
AIC
BIC
#"-1784"
"2" "3572" "3582"
#Call:
#likfit(geodata = trees4, ini.cov.pars = c(1.35, 30), lik.method = "ML")
#AIC for spatial model lower than AIC for non-spatial.
plot(tree.vario, envelope=wls.env, ylim=c(1,1.6))
lines(tree.wls, lty=2)
lines(tree.ml, lty=1)
lines(tree.wls.wave, lty=4)
errbar(tree.vario$u,tree.vario$v, yplus, yminus, add=T)
legend(100, 1.2, c("ml", "wls:exp", "wls:wave"), lty=1:3)
#
tausq (nugget)
sigmasq (sill)
phi (range)
#WLS (exp) :
1.2
52.9
19340.14
#WLS(wave)
1.25
0.26
34.77
#ML:
1.142
0.28
26.19
#******** Check to see if ML converged (also default structure for ML)
#Anisotropy—is there directionality in the degree of spatial autocorrelation
tree.vario4 = variog4(trees4, estimator.type="modulus", max.dist=140)
par(marrow=c(1,2))
plot(tree.vario, ylim=c(1,1.6))
#
plot(tree.vario4, same=T, type="b")
#Appears that autocorrelation is similar for 45,90 and 135. The degree of autocorrelation increases with #increasing
#distance in pairs that are essentially west-east?
#Model spatial autocorrelation for each direction separately:
tree.wls90 = variofit(tree.vario4$"90", ini.cov.pars=c(1,30), nugget=.5, cov.model="exponential", weights="cressie")
tree.wls135 = variofit(tree.vario4$"135", ini.cov.pars=c(1,30), nugget=.5, cov.model="exponential", weights="cressie")
tree.wls45 = variofit(tree.vario4$"45", ini.cov.pars=c(1,30), nugget=.5, cov.model="exponential", weights="cressie")
tree.wls0 = variofit(tree.vario4$"0", ini.cov.pars=c(1,30), nugget=.5, cov.model="exponential", weights="cressie")
par(mfrow=c(2,2))
plot(tree.vario4$"0", main="0")
lines(tree.wls0)
plot(tree.vario4$"45", main="45")
lines(tree.wls45)
plot(tree.vario4$"90", main="90")
lines(tree.wls90)
plot(tree.vario4$"135", main="135")
lines(tree.wls135)
# ML model which includes anisotropy
tree.ml4= likfit(trees4, ini.cov.pars=c(1,30), cov.model="exponential", lik.method="ML", fix.psiA=F, psiA=pi/4,
fix.psiR=F, psiR=2)
#The argument, psiA supplies the value (radians) for the anisotropy angle parameter.
# The argument fix.psiA indicates whether the anisotropy angle parameter is fixed (Default) or should be estimated
#(fix.psiA = F).
summary(tree.ml4)
#Summary of the parameter estimation
#----------------------------------#Estimation method: maximum likelihood
#Parameters of the mean component (trend):
# beta
#0.0646
#Parameters of the spatial component:
# correlation function: exponential
# (estimated) variance parameter sigmasq (partial sill) = 0.2743
# (estimated) cor. fct. parameter phi (range parameter) = 23.38
# anisotropy parameters:
# (estimated) anisotropy angle = 1.268 ( 73 degrees )
# (estimated) anisotropy ratio = 1.353
#Parameter of the error component:
#
(estimated) nugget = 1.138
#Transformation parameter:
# (fixed) Box-Cox parameter = 1 (no transformation)
#Practical Range with cor=0.05 for asymptotic range: 70.03544
#
#Maximised Likelihood:
# log.L n.params
AIC
BIC
# "-1734"
"6" "3480" "3510"
#non spatial model:
# log.L n.params
AIC
BIC
# "-1784"
"2" "3572" "3582"
#Call:
#likfit(geodata = trees4, ini.cov.pars = c(1, 30), fix.psiA = F,
# psiA = pi/4, fix.psiR = F, psiR = 2, cov.model = "exponential",
# lik.method = "ML")
#It appears that the model that includes anisotropy is better than the non-spatial model (AIC).
# Other options: likfit.glsm {library geoRglm} performs Monte Carlo max. likelihood in a generalized linear spatial mode
#based on a Monte Carlo sample from the conditional distribution