Population Growth Models
advertisement

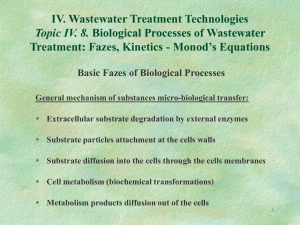
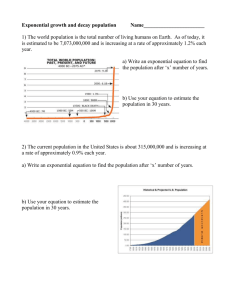
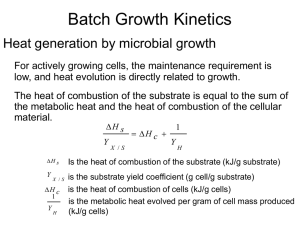
Lecture Support for Lab 2. Population Growth and Numerical Integration Population Growth Models Let’s begin with a mass balance expression for microbial biomass (X) in lakes: V dX Q X in Q X Reaction dt where Reaction refers to all of the processes relating to organism growth and death. Assuming that the flow terms can be ignored for now (a batch reactor) and that the reaction term is well described by first order kinetics, the equation becomes: V dX V k X dt and canceling the Vs: dX kX dt Here, we will explore three variation on ‘k’ (population growth models): unlimited growth (the exponential model), space-limited growth (the logistic model) and resourcelimited growth (the Monod model). Exponential or Unlimited Growth dX X dt where µ is the specific growth rate coefficient (d-1), a special case of the first order rate constant, k. Note that the units for µ could also be expressed as organisms∙organism-1∙d-1. This equation can be integrated with respect to time yielding: X t X 0 e( t ) The output from the exponential model, a J-shaped curve, and the impact of changes in the magnitude of µ, is illustrated in Figure 5-4 of Mihelcic (1999). Note, however, that after some time biomass levels predicted by the exponential model become unrealistic (2 1043 in 100 days at µ = 1 d-1). Populations do not continue to increase exponentially due to space and/or resource limitations. Logistic Growth: The Effect of Carrying Capacity Here population growth becomes limited by carrying capacity or the ability of the environment to sustain growth. We conceptualize this as space limitation (non-renewable resources, i.e density-dependent phenomena, with the effect being one of environmental resistance (Milhelcic 1999; Figure 5-5). Mathematically, the model takes the form: dX X max 1 X dt K where µmax is the maximum specific growth rate coefficient (d-1) and K is the carrying capacity (units of biomass). The analytical solution to this equation is: Xt K K X 0 max t 1 e X 0 The output from the logistic model, an S-shaped curve, is illustrated and compared with the exponential growth model in Figure 5-7 of Mihelcic (1999). Example: carrying capacity in Daphnia, Mihelcic (1999, Figure 5-6) Populations seldom, however, approach their carrying capacity because they run out of (renewable) resources first. Resource Limited Growth: The Monod Model Resource or nutrient limitation is accommodated using the Monod model: S dX max X dt Ks S where S is the nutrient (substrate, S) concentration and Ks is the half saturation constant for that nutrient, i.e. the nutrient concentration at which one-half the maximum specific growth rate is achieved. The definition of Ks and the relationship between µ and S for two values of Ks are illustrated in Mihelcic (1999, Figure 5-8). Physically, Ks reflects the affinity of enzymes for substrate (nutrients) where organisms having a low Ks value are more competitive at low nutrient levels. Example: competition in protozoa, Mihelcic (1999, Figure 5-9) Relationship to Substrate: The Yield Coefficient We can model substrate as well as biomass dynamics if we can quantify the yield of organisms per unit substrate consumed. This is accomplished through the yield coefficient: Y X S and thus the change in substrate is given by: dS 1 dX dt Y dt where the appropriate model is substituted for the dX/dt term. Respiration: The Death Coefficient Up to this point, nothing has died. We are aware that ‘death’ is going on continuously in the form of respiration which is a ‘sink’ in growth models: dX kd X dt where kd is the first order rate coefficient for endogenous respiration. Putting it All Together These models can be combined into a pair of equations useful in simulating substrate and biomass in batch culture. dX X S max 1 kd X dt K Ks S dS 1 X S max 1 kd X dt Y K Ks S This is illustrated in the classic batch growth curve (Mihelcic 1999; Figure 5-10). Consider the significance of including kd in the substrate equation. And Adding Flow For flow through systems where tributary inputs and loss to outflow are significant, we can add flow terms. dX Q Q X S X in X max 1 kd X dt V V K Ks S dS Q Q 1 X S Sin S max 1 kd X dt V V Y K Ks S where V is the reactor volume, Q is flow and Sin refers to the substrate concentration in the inflow. Numerical Integration The field of mathematical modeling was developed on the backs of those who could derive solutions for the ordinary differential equations that constituted the models. These analytical solutions have limitations, however, as described by Chapra (1997, Ch. 7): non-idealized loading functions: analytical solutions require the idealized loading functions (impulse, step, etc.) developed in Chapter 3. Actual loads behave in a much more arbitrary fashion can only be accommodated numerically. variable parameters: an analytical solution requires coefficient values remain constant over the course of the simulation. This is certainly not the case, as the specific growth rate for microbial populations, for example, may vary over the course of the simulation due to changes in environmental forcing conditions (e.g. light, temperature and nutrients). multi-segment systems: the development of 1, 2, and 3-D models for requires spatial segmentation that is inefficient to handle with analytical solutions (see Chapra 1997, p. 92, Eq. 5.21). non-linear kinetics: while linear (e.g. first order) kinetics are quite useful, there are several water quality applications that require non-linear kinetics (e.g. the Monod model). Analytical solutions cannot be obtained for these. Thus we turn to numerical methods, developing numerical or computer-based solutions. The basic idea here is the solutions are trivially simply, but exhaustively long were it not for computing power. The Euler Method In the Euler method, we transform the differential equation (here the exponential growth model) as follows: dX X dt dX X dt such that we can calculate a change in X and then add it to the original value of X to get an updated value at the end of the time interval dt: X new X old dX This approach is implemented in a computer program in the form of a loop, calculating X over an interval 0 to tmax: For t 0 to tmax dX X dt X X dX Next t The selection of a value for dt is critical to the success of the process (Spain 1982, Figure 5.1). We find as dt0, the numerical solution approaches the analytical solution. The trade-off becomes one of computation time – as we decrease the time step (dt) the solution becomes more accurate, but the number of computations that must be performed increases. Alternative numerical methods (Chapra 1997, Chapter 7: Heun’s Method, Runge-Kutta Methods) can provide accurate results at significantly longer time steps, thus reducing computation time. Output The most straightforward approach for implementing the time step is to include it explicitly in the code, e.g. For t 0 to tmax Step dt dX X dt X X dX Next t and the code would simply move along from 0 to tmax in steps of the specified dt. However, we typically wish to include an output statement in the code: For t 0 to tmax Step dt dX X dt X X dX Print t , X Next t and this would result in 1/dt lines of output – not a good idea for very small dt values. We can get around this by using a nested loop: For t 0 to tmax For k 1/ dt dX X dt X X dX Next k Print t , X Next t