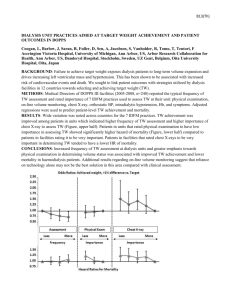
Longevity methods
advertisement

Longevity methods (replace a with the lambda character and b with the gamma!) We used 0.95 l clear plastic cages to estimate life expectancy. Food vials with standard fly medium sprinkled with dry yeast were fixed to a connector tube by a clear plastic sleeve. The flies were kept in the cages continuously at 24°C and 50% humidity in a 12 h light /12 h dark cycle until no fly remained alive. Every second day, the food vials were replaced with fresh ones, all dead flies were aspirated and sexed, and the positions of the cages relative to the lighting in the room switched. Fifteen replicate cages with equal numbers of females and males were started for each population in BOC1. In BOC2, eight replicate cages were started for each population with equal numbers of both sexes, making up 2 x 8 heterosexual milieus, and eight more with either females or males, making up 4 x 8 homosexual milieus. Unlike BOC2, dead flies were replaced with genetically marked (Ebony mutants) flies of the same sex. One homosexual cage with female descendants of Studs was accidentally lost. All cohorts were synchronously initiated for each experimental run (BOC1 and BOC2). We estimated life expectancy as the mean lifespan of the flies in each cage, with the cage being the unit of replication. Mean life span was calculated in WinModest (Pletcher 1999). Mortality parameters were estimated and hypothesis tests were performed with maximum likelihood methods implemented in WinModest. The simplest of four nested models assumes that the mortality rate at age t is µt = a ebt, with parameter a representing baseline mortality and b the rate of change in mortality with age. This, the Gompertz model, can be extended into three more complicated models by including two other mortality variables. The logistic model allows deceleration of mortality with age, denoted by s, so that µt= a ebt[1 + (a s/b)(ebt – 1)]-1, while the Gompertz-Makeham model adds age-independent mortality c, so that µt = c + a ebt . Including both s and c gives the Logistic-Makeham model, where µt=c+ a ebt[1 + (a s/b)(ebt – 1)]-1. We tested for the influence of the respective mortality parameters on differences in mortality rates between cohorts by comparing the log-likelihood when a, b or c are fixed to when they are freely estimated. Exclusion of the mortality parameter has a statistically significant effect if twice the difference in log-likelihood is larger than 3.84. We did not test for the influence of parameters unless all combinations under comparison fitted models which include that parameter. Hence, s was not tested for at all and c was ignored in BOC1. Data from the replicate cages were pooled before maximum likelihood analysis to avoid biases in the estimations of mortality parameters, which may be introduced by small sample sizes (Pletcher 1999). We censored deaths from the first check in both BOC1 and 2 to avoid inclusion of cage transfer effects in the analyses. The first mortality check in BOC1 revealed strongly elevated mortalities, which were probably associated with initiation of the cage milieus. Emergence time We estimated emergrnce time as the mean time to eclosion of the flies in each cage, the cage being the unit of replication. References Pletcher, S.D. 1999. Model fitting and hypothesis testing for age-specific mortality data. J. Evol. Biol. 12: 430–439.