General Linear Modeling - Waisman Laboratory for Brain
advertisement
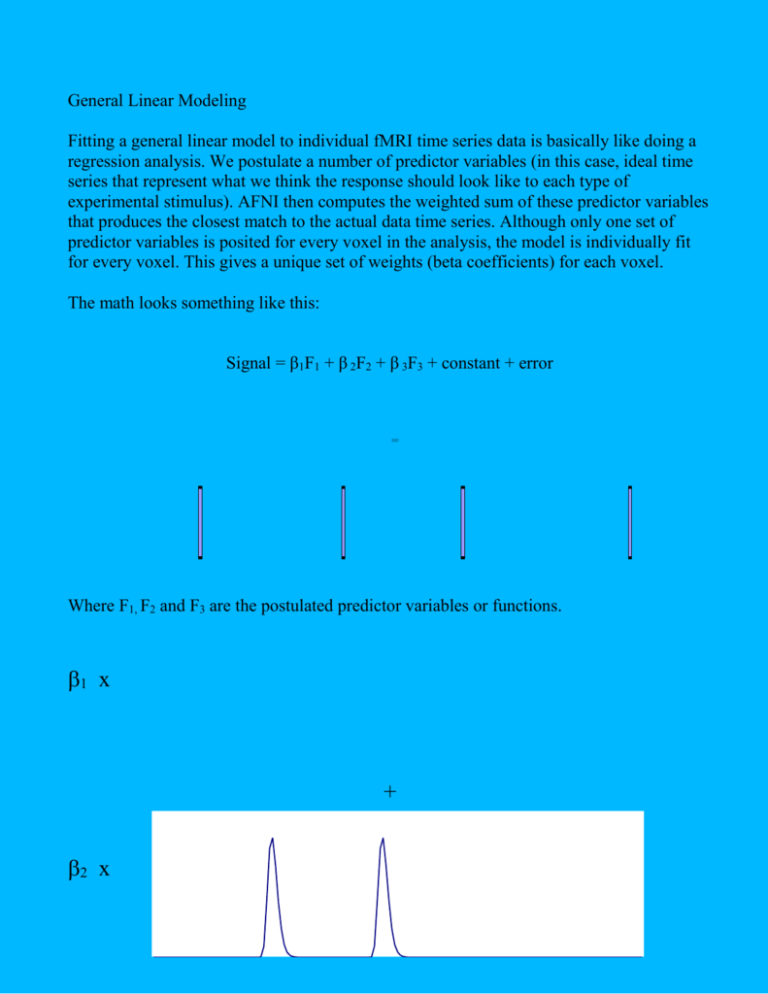
General Linear Modeling Fitting a general linear model to individual fMRI time series data is basically like doing a regression analysis. We postulate a number of predictor variables (in this case, ideal time series that represent what we think the response should look like to each type of experimental stimulus). AFNI then computes the weighted sum of these predictor variables that produces the closest match to the actual data time series. Although only one set of predictor variables is posited for every voxel in the analysis, the model is individually fit for every voxel. This gives a unique set of weights (beta coefficients) for each voxel. The math looks something like this: Signal = β1F1 + β 2F2 + β 3F3 + constant + error 00000001 Where F1, F2 and F3 are the postulated predictor variables or functions. 1 x + 2 x + 3 x = The calculated beta-weights then tell us the relative heights, or amplitudes, of the different postulated predictor functions. We can then use these calculated beta weights in two ways. First, we can compare each calculated beta weight to zero. If the beta weight differs significantly from zero for a given voxel, we may say that that voxel is activated under the experimental condition that corresponds to that beta weight. The AFNI 3dDeconvolve program reports beta weights for each postulated predictor function that you include in the model, as well as t-tests of whether each beta weight differs significantly from zero. Second, we can compare two or more beta weights to see if they are different. If two beta weights are significantly different for a given voxel, we may conclude that that voxel is more activated for one experimental condition than for another. The AFNI 3dDeconvolve program allows you to specify such comparisons between beta weights using linear contrasts. A linear contrast simply tests whether a linear combination of two or more beta weights is greater than zero. For example, to test whether β1 differs from β2 , we use the linear contrast 1 –1, which tests: H0: β1 – β2 = 0 ? i.e. is the amplitude of activation in the first condition different from that of the second condition ? Another example might be 1 –0.5 –0.5: H0: β1 – 0.5β2 – 0.5 β3 = 0 ? i.e. is the amplitude of the first condition different from the average of the activation from the second and third conditions? For each such contrast, 3dDeconvolve reports the size of the contrast, as well as the associated F-test. Multiple predictors for each condition: allowing for variable lags. It is also possible to specify a model which includes multiple predictor functions for each experimental condition. For example, we might want to allow for different lags, or latencies, in the responses to each type of stimulus. We could thus include three predictor time series for each condition – the three series would be identical, except that the ideal responses for the second and third series would be delayed in time by, e.g. 1 and 2 seconds respectively. Signal = (β1AF1A + β 2AF2A + β 3AF3A) + (β1BF1B + β 2BF2B + β 3BF3B) + constant + error So in this example, the first three terms (with the subscripts A) represent the first experimental condition), and similarly for the second three terms. With this model, the AFNI 3dDeconvolve program would compute all the beta coefficients and associated t-tests, as well as a combined test of whether the beta weights for each condition collectively differed from zero. Multiple predictors for each condition: deconvolution. Reference: Glover, G. H. (1999). Deconvolution of Impulse Response in Event-Related BOLD fMRI, NeuroImage, 9, 416 429 The extreme case of specifying multiple predictor time series for each condition is when we want to perform a deconvolution. Adopting source-filter terminology, in an event related design we can consider the fMRI time series for a single voxel as resulting from a train of initial impulse-like stimuli, which is then passed through a filter, which represents the combined effect of neural response, hemodynamic response and scanner receiver characteristics. Thus: y(t) = x(t)*h(t) where y(t) is the signal at a given voxel, x(t) is the train of input stimuli, and h(t) is the unknown filter function, that for simplicity we will call the Hemodynamic Reponse Function (HRF). The asterix denotes the convolution operation. y t = + h v x t v dv We know x(t) from our experimental design, and we have measured y(t). We should thus be able to calculate h(t) by an operation of deconvolution. In the situation where the signal is sampled discretely in time, and the HRF is assumed to have a non-zero value for only a limited time (say N seconds), this becomes: y t=² N v= 0 h v x t v To achieve this within the framework of GLM, we can model each experimental condition with a separate predictor series for each value of the index v (i.e. for each second during which the responses to stimuli in that condition are expected to be non-zero). Typically this would mean that each experimental condition would be modeled with about 16 predictor time series. Each of these time series consists of a simple string of 0s and 1s. The first series for a given condition would be have 1s corresponding to the first time point following each stimulus presentation for that condition. The second series would have 1s corresponding to the second time point following each stimulus presentation for that condition, etc. By performing the general linear model in this manner, the calculated beta weights will correspond to the estimate amplitude of the hemodynamic response 1 second, 2 seconds, 3 seconds etc following stimulus onset for each experimental condition. Example. Imagine an event-related fMRI experiment in which there is only a single event at time t = 0. Say we collect 20 seconds of data, with a TR of 1 second. Now consider a single voxel in an area which becomes activated. We thus have a 20 element vector y(t) of measured signal for that voxel: y(t) = [0 0 1 2 5 14 15 13 10 8 6 5 4 3 2 1 0 0 0 0] We also have our stimulus vector x(t) x(t) = [1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0] From our equation y t = ² N v= 0 h v x t v , we have 0 = h(0) 0 = x(1)h(0) + x(0)h(1) = h(1) 1 = x(2)h(0) + x(1)h(1) + x(0)h(2) = h(2) 2 = x(3)h(0) + x(2)h(1) + x(1)h(2) + x(0)h(3) = h(3) 5 = x(4)h(0) + x(3)h(1) + x(2)h(2) + x(1)h(3) + x(0)h(4) = h(4) and so on. So from this trivial example we can see how the function h(v) can be determined from the data. One way to look at the above analysis is that h(v) can be broken down into N separate functions, each with a non-zero value at it's corresponding time and a value of zero at all other times. Each hv(t) can then be further broken down as follows: hn(t) = bviv(t), where iv(t) = 1 whenever hv(t) is non-zero, and 0 otherwise. Bv is a scalar that represents the magnitude of hv(t) when it is non-zero (i.e. At it's specific time point). The equation then becomes: y t=² N v= 0 bv i v t x t v In experiments with more than one trial, the calculation of h(v) then involves making a least-squares estimate of the bv, where the bv represent the amplitude of h at time point v. β 1A x + β 2A x + β 3A x + β 4A x + β 5A x + β 6A x + β 7A x + β 8A x + β 1B x + β 2B x + β 3B x + β 4B x + β 5B x + β 6B x + β 7B x + β 8B x + constant + error = signal The beta weights give us: β 1A - β 8A β 1B - β 8B








