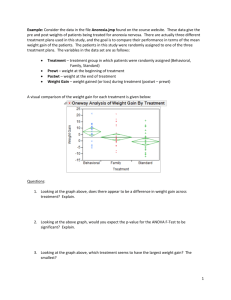
One-way ANOVA
advertisement

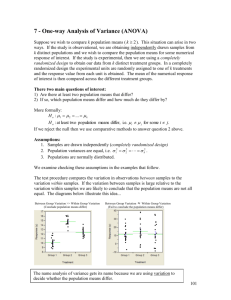
One-way Analysis of Variance (ANOVA) (Chapter 8, Daniel) Suppose we wish to compare k population means ( k 2 ). This situation can arise in two ways. If the study is observational, we are obtaining independently drawn samples from k distinct populations and we wish to compare the population means for some numerical response of interest. If the study is experimental, then we are using a completely randomized design to obtain our data from k distinct treatment groups. In a completely randomized design the experimental units are randomly assigned to one of k treatments and the response value from each unit is obtained. The mean of the numerical response of interest is then compared across the different treatment groups. There two main questions of interest: 1) Are there at least two population means that differ? 2) If so, which population means differ and how much do they differ by? More formally: H o : 1 2 ... k H a : at least two population means differ, i.e. i j for some i j. If we reject the null then we use comparative methods to answer question 2 above. Assumptions: 1. Samples are drawn independently (completely randomized design) 2. Population variances are equal, i.e. 12 22 k2 . 3. Populations are normally distributed. We examine checking these assumptions in the examples that follow. The test procedure compares the variation in observations between samples to the variation within samples. If the variation between samples is large relative to the variation within samples we are likely to conclude that the population means are not all equal. The diagrams below illustrate this idea... Between Group Variation >> Within Group Variation (Conclude population means differ) Between Group Variation Within Group Variation (Fail to conclude the population means differ) The name analysis of variance gets its name because we are using variation to decide whether the population means differ. 1 Within Group Variation To measure the variation within groups we use the following formula: (n1 1) s12 (n2 1) s22 (nk 1) sk2 s n1 n2 nk k 2 W Note: This is an extension of pooled variance estimate from the two-sample t-Test 2 for independent samples ( s p ) to more than two populations. This is an estimate of the variance common to all k populations ( 2 ). Between Group Variation To measure the variation between groups we use the following formula: n1 ( x1 x ) 2 n2 ( x2 x ) 2 nk ( xk x ) 2 s k 1 where, 2 B xi sample mean response for the i th treatment group x grand mean of the response from all treatment groups The larger the differences between the k sample means for the treatment groups are, the larger s B2 will be. If the null hypothesis is true, i.e. the k treatments have the same population mean, we expect the between group variation to the same as the within group variation ( s B2 sW2 ). If s B2 sW2 we have evidence against the null hypothesis in support of the alternative, i.e. we have evidence to suggest that at least two of the population means are different. Test Statistic F n = overall sample size s B2 ~ F-distribution with numerator df = k -1 and denominator df = n – k sW2 The larger F is the strong the evidence against the null hypothesis, i.e. Big FReject Ho. 2 EXAMPLE 1 - Weight Gain in Anorexia Patients Data File: Anorexia.JMP These data give the pre- and post-weights of patients being treated for anorexia nervosa. There are actually three different treatment plans being used in this study, and we wish to compare their performance in terms of the mean weight gain of the patients. The patients in this study were randomly assigned to one of three therapies. The variables in the data file are: group - treatment group (1 = Family therapy, 2 = Standard therapy, 3 = Cognitive Behavioral therapy) Treatment – treatment group by name (Behavioral, Family, Standard) prewt - weight at the beginning of treatment postwt - weight at the end of the study period Weight Gain - weight gained (or lost) during treatment (postwt-prewt) We begin our analysis by examining comparative displays for the weight gained across the three treatment methods. To do this select Fit Y by X from the Analyze menu and place the grouping variable, Treatment, in the X box and place the response, Weight Gain, in the Y box and click OK. Here boxplots, mean diamonds, normal quantile plots, and comparison circles have been added. Things to consider from this graphical display: Do there appear to be differences in the mean weight gain? Are the weight changes normally distributed? Is the variation in weight gain equal across therapies? 3 Checking the Equality of Variance Assumption To test whether it is reasonable to assume the population variances are equal for these three therapies select UnEqual Variances from the Oneway Analysis pull down-menu. Equality of Variance Test H o : 12 22 k2 H a : the population variances are not all equal We have no evidence to conclude that the variances/standard deviations of the weight gains for the different treatment programs differ (p >> .05). ONE-WAY ANOVA TEST FOR COMPARING THE THERAPY MEANS To test the null hypothesis that the mean weight gain is the same for each of the therapy methods we will perform the standard one-way ANOVA test. To do this in JMP select Means, Anova/t-Test from the Oneway Analysis pull-down menu. The results of the test are shown in the Analysis of Variance box. s B2 307.22 sW2 56.68 The between group variation is 5.42 times larger than the within group variation! This provides strong evidence against the null hypothesis (p = .0065). The p-value contained in the ANOVA table is .0065, thus we reject the null hypothesis at the .05 level and conclude statistically significant differences in the mean weight gain experienced by patients in the different therapy groups exist. MULTIPLE COMPARISONS Because we have concluded that the mean weight gains across treatment method are not all equal it is natural to ask the secondary question: Which means are significantly different from one another? We could consider performing a series of two-sample t-Tests and constructing confidence intervals for independent samples to compare all possible pairs of means, however if the number of treatment groups is large we will almost certainly find two treatment means as being significantly different. Why? Consider a situation where we have k = 7 different treatments that we wish to compare. To compare all possible pairs of means (1 vs. 2, 1 k (k 1) 21 two-sample t-Tests. If vs. 3, …, 6 vs. 7) would require performing a total of 2 we used P(Type I Error) .05 for each test we expect to make 21(.05) 1 Type I Error, i.e. we expect to find one pair of means as being significantly different when in fact they are not. This problem only becomes worse as the number of groups, k , gets larger. 4 Experiment-wise Error Rate Another way to think about this is to consider the probability of making no Type I Errors when making our pair-wise comparisons. When k = 7 for example, the probability of making no Type I Errors is (.95) 21 .3406 , i.e. the probability that we make at least one Type I Error is therefore .6596 or a 65.96% chance. Certainly this unacceptable! Why would conduct a statistical analysis when you know that you have a 66% of making an error in your conclusions? This probability is called the experiment-wise error rate. Bonferroni Correction There are several different ways to control the experiment-wise error rate. One of the easiest ways to control experiment-wise error rate is use the Bonferroni Correction. If we plan on making m comparisons or conducting m significance tests the Bonferroni Correction is to simply use as our significance level rather than . This simple m correction guarantees that our experiment-wise error rate will be no larger than . This correction implies that our p-values will have to be less than rather than to be m considered statistically significant. Multiple Comparison Procedures for Pair-wise Comparisons of k Population Means When performing pair-wise comparison of population means in ANOVA there are several different methods that can be employed. These methods depend on the types of pair-wise comparisons we wish to perform. The different types available in JMP are summarized briefly below: Compare each pair using the usual two-sample t-Test for independent samples. This choice does not provide any experiment-wise error rate protection! (DON’T USE) Compare all pairs using Tukey’s Honest Significant Difference (HSD) approach. This is best choice if you are interested comparing each possible pair of treatments. Compare with the means to the “Best” using Hsu’s method. The best mean can either be the minimum (if smaller is better for the response) or maximum (if bigger is better for the response). Compare each mean to a control group using Dunnett’s method. Compares each treatment mean to a control group only. You must identify the control group in JMP by clicking on an observation in your comparative plot corresponding to the control group before selecting this option. 5 Multiple Comparison Options in JMP EXAMPLE 1 - Weight Gain in Anorexia Patients (cont’d) For these data we are probably interested in comparing each of the treatments to one another. For this we will use Tukey’s multiple comparison procedure for comparing all pairs of population means. Select Compare Means from the Oneway Analysis menu and highlight the All Pairs, Tukey HSD option. Beside the graph you will now notice there are circles plotted. There is one circle for each group and each circle is centered at the mean for the corresponding group. The size of the circles are inversely proportional to the sample size, thus larger circles will drawn for groups with smaller sample sizes. These circles are called comparison circles and can be used to see which pairs of means are significantly different from each other. To do this, click on the circle for one of the treatments. Notice that the treatment group selected will be appear in the plot window and the circle will become red & bold. The means that are significantly different from the treatment group selected will have circles that are gray. These color differences will also be conveyed in the group labels on the horizontal axis. In the plot below we have selected Standard treatment group. 6 The results of the pair-wise comparisons are also contained the output window. The matrix labeled Comparison for all pairs using Tukey-Kramer HSD identifies pairs of means that are significantly different using positive entries in this matrix. Here we see only treatments 2 and 3 significantly differ. The next table conveys the same information by using different letters to represent populations that have significantly different means. Notice treatments 2 and 3 are not connected by the same letter so they are significantly different. Finally the CI’s in the Ordered Differences section give estimates for the differences in the population means. Here we see that mean weight gain for patients in treatment 3 is estimated to be between 2.09 lbs. and 13.34 lbs. larger than the mean weight gain for patients receiving treatment 2 (see highlighted section below). 7 EXAMPLE 2 – Motor Skills in Children and Lead Exposure These data come from a study of children living in close proximity to a lead smelter in El Paso, TX. A study was conducted in 1972-73 where children living near the lead smelter were sampled and their blood lead levels were measured in both 1972 and in1973. IQ tests and motor skills tests were conducted on these children as well. In this example we will examine difference in finger-wrist tap scores for three groups of children. The groups were determined using their blood lead levels from 1972 and 1973 as follows: Lead Group 1 = the children in this group had blood levels below 40 micrograms/dl in both 1972 and 1973 (we can think of these children as a “control group”). Lead Group 2 = the children in this group had lead levels above 40 micrograms/dl in 1973 (we can think of these children as the “currently exposed” group). Lead Group 3 = the children in this group had lead levels above 40 microgram/dl in 1972, but had levels below 40 in 1973 (we can think of these children as the “previously exposed” group). The response that was measured (MAXFWT) was the maximum finger-wrist tapping score for the child using both their left and right hands to do the tapping (to remove hand dominance). These data are contained in the JMP file: Maxfwt Lead.JMP. Select Fit Y by X and place Lead Group in the X,Factor box and MAXFWT in the Y,Response box. After selecting Quantiles and Normal Quantile Plots we obtain the following comparative display. The wrist-tap scores appear to be approximately normally distributed. The variance in the wrist-tap scores may appear to be a bit larger for Lead Group 1, however because this is the largest group we expect to see more observations in the extremes. The interquartile ranges for the three lead groups appear to be approximately equal. We can check the equality of variance assumption formally by selecting the UnEqual Variance option. 8 Formally Checking the Equality of the Population Variances None of the equality of variance tests suggests that the population variances are unequal (p > .05). ONE-WAY ANOVA FOR COMPARING THE MEAN WRIST-TAP SCORES ACROSS LEAD GROUP To test the null hypothesis that the mean finger wrist-tap score is the same for each of the lead exposure groups we will perform the standard one-way ANOVA test. To do this in JMP select Means, Anova from the Oneway Analysis pull-down menu. The results of the test are shown in the Analysis of Variance box. The p-value contained in the ANOVA table is .0125, thus we reject the null hypothesis at the .05 level and conclude that statistically significant differences in the mean finger wrist tap scores of the children in the different lead exposure groups exist (p < .05). Multiple Comparisons using Tukey’s HSD Only the mean finger wrist-tap scores of lead groups 1 and 2 significantly differ, i.e. children who currently have high lead levels have a significantly smaller mean than the children who did not have high lead levels in either year of the study. 9 Tukey’s HSD Pair-wise Comparisons (cont’d) The results of the pair-wise comparisons are also contained the output window. The matrix labeled Comparison for all pairs using Tukey-Kramer HSD identifies pairs of means that are significantly different using positive entries in this matrix. Here we see only lead groups 1 and 2 significantly differ. The next table conveys the same information by using different letters to represent populations that have significantly different means. Notice lead groups 1 and 2 are not connected by the same letter so they are significantly different. Finally the CI’s in the Ordered Differences section give estimates for the differences in the population means. Here we see that the mean finger wrist tap score for children who currently have a high blood lead level is estimated to be between .83 and 14.18 taps smaller than the mean finger wrist tap score for children who did not have high lead levels in either year of the study. 10 Randomized Complete Block (RCB) Designs (Section 8.3 Daniel) EXAMPLE 1 – Comparing Methods of Determining Blood Serum Level Data File: Serum-Meth.JMP The goal of this study was determine if four different methods for determining blood serum levels significantly differ in terms of the readings they give. Suppose we plan to have 6 readings from each method which we will then use to make our comparisons. One approach we could take would be to find 24 volunteers and randomly allocate six subjects to each method and compare the readings obtained using the four methods. (Note: this is called a completely randomized design). There is one major problem with this approach, what is it? Instead of taking this approach it would clearly be better to use each method on the same subject. This removes subject to subject variation from the results and will allow us to get a clearer picture of the actual differences in the methods. Also if we truly only wish to have 6 readings for each method, this approach will only require the use of 6 subjects versus the 24 subjects the completely randomized approach discussed above requires, thus reducing the “cost” of the experiment. The experimental design where each patient’s serum level is determined using each method is called a randomized complete block (RCB) design. Here the patients serve as the blocks; the term randomized refers to the fact that the methods will be applied to the patients blood sample in a random order, and complete refers to the fact that each method is used on each patients blood sample. In some experiments where blocking is used it is not possible to apply each treatment to each block resulting in what is called an incomplete block design. These are less common and we will not discuss them in this class. The table below contains the raw data from the RCB experiment to compare the serum determination methods. Method 2 3 4 Subject 1 1 360 435 391 502 2 1035 1152 1002 1230 3 632 750 591 804 4 581 703 583 790 5 463 520 471 502 6 1131 1340 1144 1300 11 Visualizing the need for Blocking Select Fit Y by X from the Analyze menu and place Serum Level in the Y, Response box and Method in the X, Factor box. The resulting comparative plot is shown below. Does there appear to be any differences in the serum levels obtained from the four methods? This plot completely ignores the fact that the same six blood samples were used for each method. We can incorporate this fact visually by selecting Oneway Analysis > Matching Column... > then highlight Patient in the list. This will have the following effect on the plot. Now we can clearly see that ignoring the fact the same six blood samples were used for each method is a big mistake! On the next page we will show how to correctly analyze these data. 12 Correct Analysis of RCB Design Data in JMP First select Fit Y by X from the Analyze menu and place Serum Level in the Y, Response box, Method in the X, Factor box, and Patient in the Block box. The results from JMP are shown below. Notice the Y axis is “Serum Level – Block Centered”. This means that what we are seeing in the display is the differences in the serum level readings adjusting for the fact that the readings for each method came from the same 6 patients. Examining the data in this way we can clearly see that the methods differ in the serum level reported when measuring blood samples from the same patient. The results of the ANOVA clearly show we have strong evidence that the four methods do not give the same readings when measuring the same blood sample (p < .0001). The tables below give the block corrected mean for each method and the block means used to make the adjustment. As was the case with one-way ANOVA (completely randomized) we may still wish to determine which methods have significantly different mean serum level readings when measuring the same blood sample. We can again use multiple comparison procedures. 13 Select Compare Means... > All Pairs, Tukey’s HSD. We can see that methods 4 & 2 differ significantly from methods 1 & 3 but not each other. The same can be said for methods 1 & 3 when compared to methods 4 & 2. The confidence intervals quantify the size of the difference we can expect on average when measuring the same blood samples. For example, we see that method 4 will give between 90.28 and 255.05 higher serum levels than method 3 on average when measuring the same blood sample. Other comparisons can be interpreted in similar fashion. 14