based methods in the environment and hospital patients
advertisement

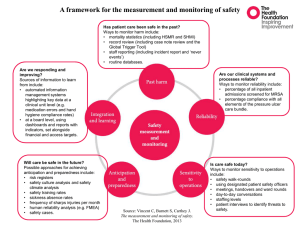
Rapid and sensitive detection of MRSA by PCR-based methods in the environment and hospital patients. by Dr. Corinne Whitby Department of Biological Sciences, Wivenhoe Park, University of Essex, Colchester, Essex, CO4 3SQ. Tel: +44 (0) 1206 872062, Fax: +44 (0) 1206 872592. Email: cwhitby@essex.ac.uk. MRSA ‘the super bug’ Methicillin-resistant Staphylococcus aureus (MRSA) was first discovered in the UK in 1961. It is a major pathogen associated with nosocomial disease both in the community and in hospitals. The number of MRSA infections is increasing with >1,600 deaths reported in 2006 in the UK alone. SEM of MRSA SEM of MRSA Screening, treatments and prevention Screening- by taking nasal swabs upon arrival at hospital. Treatments- Vancomycin & teicoplanin antibiotics- but resistant strains have been discovered. Additional antibiotics are used to treat these severe infections. Maggot therapy- to treat necrotic tissue from MRSA infections. Phage therapy- 95% efficacy against Staphylococcus isolates. J. Infect. Dis. (2003). 187 (4): 613–24. New antibiotics are currently in clinical trials. e.g. Ceftaroline. Prevention- handwashing, essential oils e.g. Tea tree oil Ruptured MRSA abscess Genetics of MRSA Methicillin resistance is mediated by the mecA gene encoding for penicillin-binding protein 2a (PBP-2a) & allows S. aureus to be oxacillin resistant. Mueller Hinton agar showing MRSA resistant to oxacillin disk. mecA gene is located on a mobile genetic element called the staphylococcal cassette chromosome (SCC). Expression of PBP-2a is controlled by mecR1 & mecI regulator genes located upstream of mecA gene. Isolates with mutations in the mec regulators may phenotypically be highly resistant to methicillin & so are also clinically relevant. SCCmecII is most abundant in hospitals, whereas SCCmecIV is most abundant community-acquired MRSA. Nature Reviews (2009) 7: 629 Diagnostic identification of MRSA Traditional methods require culturing, phenotypic and biochemical characterisation which is time-consuming. Molecular methods e.g. Gene probing, DNA sequencing of rRNA genes, FISH and PCR have all been used- but may not differentiate between MSSA and MRSA. Other methods include Real-time Q-PCR and MLST. A PCR assay which detects polymorphisms in the 16S-23S rRNA internal transcribed spacer region (ITS) may provide a rapid alternative for identifying S. aureus spp. Our Hypothesis: In hospitals MRSA spreads between the patient and the environment and vice versa and it is likely that identical MRSA genotypes colonise both patient and environment. Our Aims: • To identify MRSA in a hospital ITU, directly from the environment without culturing & determine how the environment may act as a reservoir for MRSA. • To apply ITS-PCR to differentiate between MRSA and MSSA isolates from both patient and the environment. • To analyse the genetic diversity of mecA, mec regulator genes in MRSA isolates cultured from both clinical and environmental samples. Methods/ Approaches Sampling hospital environment Direct DNA extraction Clinical samples Isolation & Screening of MRSA isolates Antimicrobial susceptibility testing PCR amplification of ITS region PAGE analysis of ITS region DNA extraction PCR amplification of mecA and mec regulator genes mecA gene sequencing, RFLP analysis of mec regulator genes Results 110 environmental samples were taken from hospital surfaces e.g. bed mattresses, floors, walls, desktops, beds, computer keyboards, ventilator equipment and trolleys. • Only 7 isolates were obtained from the environment and all were MRSA (determined by PCR amplification of mecA gene and culture). • 60 isolates were obtained from clinical samples from patients e.g. throat swab, sputum, blood culture, nose swab, wound swab, abscess, trachaeal, hip fluid. • 45% of the clinical isolates were MRSA (determined by PCR amplification of mecA gene and culture). PAGE analysis of ITS-PCR. PAGE is a high resolution method for fractionating DNA. It allows rapid, high throughput screening of samples. PAGE analysis of ITS-PCR from 60 clinical isolates. 400 bp 300 bp ITS-type 6 is predominant. 200 bp 1 6 6 7 7 1516 6 6 8 9 10 6 11 9 9 12 6 7 6 8 6 6 6 13 13 6 2 ITS-type 100 bp 400 bp 300 bp 200 bp 100 bp ITS type 1: EMRSA-16 NCTC13143. ITS type 2: MSSA NCTC6571 PAGE analysis of ITS-PCR from 7 environmental MRSA isolates. ITS-type 6 also detected in 1 environmental isolate. ITS-type 3,4 and 5 were unique to environmental isolates i.e. not found in the clinical isolates suggesting different genotypes were colonizing the environment compared to the patient. ITS type 2: MSSA NCTC6571 400 bp 300 bp 200 bp 3 3 3 4 5 6 2 2 100 bp Example PAGE of ITS-types directly from 110 hospital environmental samples without culturing. 400 bp 300 bp ITS-type 6 also found in several environmental samples. 200 bp 100 bp 400 bp 300 bp 200 bp 100 bp ITS type 1: EMRSA16 NCTC13143. ITS type 2: MSSA NCTC6571 Distribution of ITS-types from S. aureus isolates (both environmental and clinical) that were mecA+ or mecA-. Total number of isolates 35 30 25 20 Number of mecA- 15 Number of mecA+ 10 5 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 ITS subtype 17 different ITS profiles were identified: 6 patterns for MRSA, 7 patterns for MSSA, and 4 patterns for both MSSA and MRSA. 22 isolates that were mecA+ were from ITS type 6. Analysis of mecA and mec regulatory genes (mecI, mecRI). mecA gene PCR amplification: No mecA PCR products were obtained from the environment but mecA PCR products obtained for all 7 environmental & 27 clinical isolates. mecI gene PCR amplification: No environmental isolates generated mecI PCR products. M C206 MseI C224 M MseI Uncut Uncut M C206 C224 2 clinical isolates (C206, C224) generated mecI PCR products and were MRSA. Restriction digestion with MseI generated bands of 350, 130 bp, and <30bpconsistent with other workers. mecRI PCR amplification: 2 environmental & 17 clinical isolates generated mecRI PCR products. All were MRSA except 1 clinical isolate which was MSSA. 600 bp 400 bp 200 bp PCR amplification and restriction digest of mecI genes from 2 clinical S. aureus isolates. Conclusions I • ITS-PCR was successfully used to directly detect S. aureus spp. from hospital environments without culture and may be applied for rapid screening. • 56% of isolates were MRSA & represented ITS- types 1,3,4,5,16 & 17. Screen for these? • ITS-type 6 predominated clinical samples with MRSA representatives found in both environmental isolates and environmental samples. Screen for these also? • Different ITS-types were colonizing the environment compared to the patient and were MRSA. Thus despite stringent disinfection surfaces are a reservoir for MRSA which have the potential to transfer to the patient. • No isolate had ITS-profiles identical to the control strain EMRSA-16, a common organism recovered from hospitals in the UK- these S. aureus spp. may have arisen from multiple external sources. Conclusions II • No mecA PCR products were obtained directly from the environment- MRSA was below detection limits. A ‘nested’ PCR amplification approach may improve sensitivity? • mecA PCR products were obtained for all 7 environmental & 27 clinical isolates and corroborated the antibiotic susceptibility testing. • BLASTN analysis of the mecA sequences from clinical and environmental isolates had 98-100% identity to mecA genes from S. aureus spp. • Only two clinical isolates (C206, C224) generated mecI PCR products. Both were MRSA. No environmental isolates generated mecI PCR products. • mecRI gene was detected in 19 isolates (18 of which were MRSA). • It is possible that MRSA strains only have mecA and acquire the mec regulator genes later. • Large genetic diversity obtained with all S. aureus isolates suggest there is a broad-range of MRSA clones in the hospital and gene transfer may occur whereby mecA homologues may be acquired from the environmental reservoir. Acknowledgements We thank: Hospital Infection Society, Santander University of Essex for help with funding this work.