ChIP-seq Methods & Analysis_pt1
advertisement

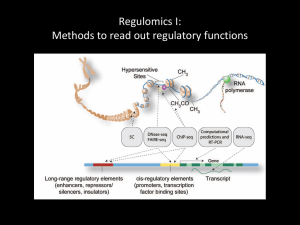
ChIP-seq Methods & Analysis Gavin Schnitzler Asst. Prof. Medicine TUSM, Investigator at MCRI, TMC gschnitzler@tuftsmedicalcenter.org 617-636-0615 What is CBI? • The Computational Biology Initiative (CBI) is a forum for Tufts researchers to collaborate and develop competitive grants in ‘omics research. • Beta Site: sites.tufts.edu/cbi Founding CBI Members: Lax Iyer, Tufts/TMC,Please Peter Castaldi, Larry RegisterTMC, at the webParnell, site: HNRCA, Gordon Huggins, TMC, Gavin Schnitzler, TMC, Joshua Ainsley, Tufts, Lionel Zupan, http://tinyurl.com/CBISymposium Tufts For more information contact: lax.iyer@tufts.edu CBI Partners: TUSM Provost’s office: Tufts Collaborates! and Tufts Innovates! Grant Awards What does CBI do? • Bring together experts in computational biology, genetics & genomics across all Tufts campuses. • Help create & maintain computational biology resources • Raise awareness and educate researchers in genomics How can we work together? • Discuss your research projects with us • Attend Symposia/talks/courses & send ideas for new ones • Contribute to our website: Your ideas, protocols/how-tos • Attend an open meeting of CBI to discuss how we could work together and what needs to be done! • Easiest way to contact: lax.iyer@tufts.edu ChIP-seq COURSE OUTLINE • Day 1: ChIP techniques, library production, USCS browser tracks • Day 2: QC on reads, Mapping binding site peaks, examining read density maps. • Day 3: Analyzing peaks in relation to genomic feature, etc. • Day 4: Analyzing peaks for transcription factor binding site consensus sequences. • Day 5: Variants & advanced approaches. ChIP-seq big picture • Combine “Next-Generation” sequencing with Chromatin Immunoprecipitation to identify genomewide chromatin binding sites. • Select (and identify) fragments of DNA that interact with specific proteins such as: – Transcription factors – Modified histones – RNA Polymerase (survey actively transcribe portions of the genome) – DNA polymerase (investigate DNA replication) – DNA repair enzymes – Or fragments of DNA that are modified: e.g. CpG methylation DAY 1 LECTURE OUTLINE • ChIP method, validating your antibody. • Next Generation Sequencing technology • Preparing a ChIP-seq library • Choosing options for sequencing • Looking at published ChIP-seq results -Exercise: Tracks on the UCSC browser Getting ready for a ChIP experiment First, validate your antibody for ChIP Step 1: Show reactivity on a Western (denatured epitope). Requirements: Rough quantification, true band should be >50% of combined signal from other bands (ENCODE guidelines) Step 2: Show IP ability. Perform IP, run Western and reprobe with same Ab (IP-Western) Show that a control antibody, non-immune IgG or to an irrelevant protein does not IP your protein in IP-Western. Chromatin Immunoprecipitation (ChIP) Step 1: Cross-linking • Treat cells with formaldehyde: protein-protein and protein-DNA cross-links stop transcription factors in their tracks (DAY 0) Cross-linking can be done on: •Suspension cells •Adherent Cells •Tissues •Anatomical structures Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Formaldehyde Crosslinking DNA-DNA Protein-Protein Other Options Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham ChIP protocol--Day 1 •Fix cells or tissues immediately (formaldehyde most frequently used) to ‘freeze’ protein:DNA interactions in place. •Stop the cross-linking reaction (glycine, for formaldehyde). •[if using whole tissues, will want to grind them up with a Tissue Disruptor, or similar small stick blender, & pellet the resulting tissue fragments] •Solubilize cells or tissue fragments in buffer with detergents (NP40, TX100, Tween and/or SDS). Should also contain protease inhibitors. •Sonicate to break up chromatin into manageable sizes (for some applications you might use micrococcal nuclease digestion). Probe sonicators vs. Bath sonicators + Every department has one. -Requires practice to use -Hard to prevent frothing -Variable results (day to day or experimenter to experimenter) + Easy to use + No frothing + Consistent results. -Rare. Not easy to get access to one. -Very expensive ($40k-$50k for a “Bioruptor”) Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Check Sonication 31 pulses 21 pulses 11 pulses 6 pulses Sonication results in many different sized fragments and should be optimized for your system 10 kbp 3 kbp 500 bp 100 bp Ideal size range will generally have peak EtBr intensity between ~300 and ~500 bp. Can do quick check with un-crosslinked chromatin, but for accurate size range need to reverse cross-links first. Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Antibodies & Resins for ChIP One Step • 1º Ab to protein of interest & control Ab • Recover complexes on protein A or protein G beads (overnight) (A+G OK for antibodies from mouse, A for antibodies from rabbit, check specificity for your Ab before beginning). • Wash to remove unbound protein Two steps (if your Ab binds weakly to protein A/G) • 1º Ab & control (first incubation) • 2º Ab (e.g. rabbit antigoat) • Recover complexes on protein A beads • Wash to remove unbound protein ChIP reactions Positive Antibody: e.g. mouse antiPolII primary with rabbit antimouse IgG secondary Negative Control: Nonspecific Rabbit IgG with rabbit antimouse IgG secondary 2 ° 1° 2 ° 1° ctrl • Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Wash with buffer containing nonionic detergent (DAY 2) Washes •Warning! Washes of Protein A or Protein G agarose beads by centrifugation results in great loss. •Protein A or G attached to magnetic beads much better. Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Elution & Crosslink Reversal 1) Addition of NaHCO3 causes antibodies to release from their target proteins, and incubation at 65º for 6 hours or more reverses crosslinks. DNA fragments are now free in solution . 2) [Can treat with proteinase K (to remove any remaining protein & RNase A to remove any residual RNA - not always necessary] 3) Column-based PCR Purification kit to purify DNA. [Read product literature & be mindful of kit limitations, e.g. standard Qiagen kit is poor at recovering short (<100 bp) or long (>2kb) fragments. This is actually good for some applications, like ChIP-seq, since it removes small fragments that may contribute the majority of ends to your libraries, but could cause problems for others] A standard ChIP protocol from cultured cells: Chromatin immunoprecipitation (ChIP). Carey MF, Peterson CL, Smale ST. Cold Spring Harb Protoc. 2009 Sep;2009(9):pdb.prot5279. doi: 10.1101/pdb.prot5279. PMID:20147264 Target Primers 1) Ratio of: (PolII ChIP)/(input) with Target Gene Primers, should be much greater than (PolII ChIP)/(input) with Control Region Primers (shows enrichment at target site). 2) Ratio of (PolII ChIP)/(IgG ChIP) for Target Gene Primers should be much greater than (PolII ChIP)/(IgG ChIP) for control region primers (controls for non-specific pull down). MW 1 ul 0.2% Input 1 ul 0.02% Input 1 ul 0.004% Input 1 ul IgG ChIP 1 ul polII ChIP 1 ul 0.2% Input 1 ul 0.02% Input 1 ul 0.004% Input 1 ul IgG ChIP 1 ul polII ChIP MW Old school PCR to Verify ChIP Control Region Primers 10% Input diluted into 50 ml = 0.2% Input/ml • Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham Quantitative PCR to Verify ChIP 1) Convert Ct values into ‘approx. relative template concentration’ values by taking 1.9^-Ct. 2) [IP@target]/[input@target] / [IP@control]/[input@/control], gives fold-enrichment at target locus vs. control * 3) [IP@target]/[Ctrl_Ab@target] / [IP@control]/[Ctrl_Ab@conrol], gives indication of antibody specificity and effectiveness of washes. 4) Both should be high before proceeding w/ ChIP-seq. * If you don’t already know positive control & negative control target loci, for your Ab & your cells then: a) Comb through the literature for prior studies using antibodies to your protein of interest & similar systems. Design a few primer sets, because transcription factor binding sites can differ greatly between cell types, but a few are likley to be the same. b) Alternatively, use mRNA expression data, etc. to identify candidate loci at which the FOI might bind & try a bunch, along with predicted negative controls (e.g. 20kb down from the end of the candidate gene). A high # in comparisons 2 & 3 above will confirm your ChIP effectiveness & identify + & - control regions to use in later ChIPs. For this sort of search a larger sonication size (~700 bp) Cluster Account Test Break • Find Putty.exe on the desktop & launch • Set up connection to cluster.uit.tufts.edu • Login w/ tufts UserID & password. Accessing the cluster from your own computer For Windows machines, get Putty at: www.putty.org For MACs, open the “terminal” utility & type: ssh tuftsid@cluster.uit.tufts.edu DAY 1 LECTURE OUTLINE • ChIP method, validating your antibody. • Next Generation Sequencing technology • Preparing a ChIP-seq library • Choosing options for sequencing • Looking at published ChIP-seq results -Exercise: Tracks on the UCSC browser ChIP-chip (ChIP2) “the pre-sequencing technology” -Limited to organisms with available genomic microarrays (or you’ll need to make your own) -Microarrays with oligos covering whole mammalian genomes are very expensive (many arrays per sample) ChIP Input WGA -Can be economical for model organisms with small genomes & commercially available arrays (or for limited analysis: e.g. promoter regions). Whole genome amplification (WGA) allows good probe signal from small starting samples. -Subject to hybridization curve limitations & hyb. artifacts Label w/ Cy Dyes Apply to microarray Compare Red/Green signal intensities to identify binding sites Adapted from ChIP Workshop by Charlie Nicolet, Heather N. Witt & Peggy Farnham ChIP-seq Immunoprecipitate POI= protein of interest High-throughput sequencing of DNA ends Map sequence tags to genome & identify peaks Prepare sequencing library Release DNA Adapted from slide set by: Stuart M. Brown, Ph.D., Center for Health Informatics & Bioinformatics, NYU School of Medicine ChIP-Seq advantages • Doesn’t require a specially-constructed microarray • Works with any sequenced genome (better if it’s also well annotated) • Can be economical: • At ~160 Million reads, one lane can give you all the binding sites in the genome •Multiplexing can allow multiple samples per lane Limitations: • Like arrays, can’t make sense of repeat regions. • Always genomewide: …which is great for transcription factor binding sites & some histone modifications (where only a few places in the genome have many reads, over a low reads/kb background) … can be problematic for very common events like nucleosome positions & CpG methylation (where most places in genome have roughly equal reads/kb, thus 160M reads still gives read # at any one locus that is too low to quantitate). What is next generation sequencing? Next generation = the stuff that came after standard one template per reaction Sanger sequencing Next generation sequencing(NGS) = High throughput sequencing(HTS) = Deep sequencing It’s all just massively parallel sequencing. HTS Commonalities (so far) Fragmentation of starting DNA Ligation with custom adapters Library amplification on a solid surface (either bead or glass) Direct detection of each incorporated nucleotide Hundreds of thousands to billions of reactions Shorter read lengths than capillary sequencers Count based data for quantitation Sampling both ends of every fragment sequenced (paired end reads) Sequencing Platforms Company Platform Amplification Roche emPCR Illumina 454 Illumina/ Solexa Bridge PCR Life SOLiD emPCR Sequencing Synthesis, Pyrosequencing Synthesis, Fluorescence Ligation, Fluorescence emPCR Synthesis, H+ detection Life Ion Torrent PacBio RS None Synthesis, ZMW fluorescence Oxford Nanopore ION None? Nanopore current flow To learn all about these cool upcoming technologies, check out Josh Ainsley’s 1st day RNA seq course slides at: http://sites.tufts.edu/cbi/resources/rna-seq-course/lectures/ Illumina Sequencing (the current mostused standard) Sequencing-bysynthesis Separate fluorescent tags on each nucleotide Reversible terminators Library preparation steps Fragmentation End repair and A-tailing Adapter ligation Amplification gives distinct ends Getting your library on a flowcell Cluster generation – Bridge amplification Cluster generation – Bridge amplification Sequencing Single end – sequence one end Paired end – sequence both ends (separate runs w/ primer for orange end, & then w/ primer for green) TruSeq Adapters Forked adapters Not complete without PCR Indexes/ barcodes allow for multiplex sequencing using a third sequencing read (currently up to 24) In-line multiplex adapters Index/barcode is in the first 4-8 bases of the sequencing read Indexes/barcodes allow for multiplex sequencing Up to 24 separate libraries on the same Illumina HiSeq lane. After sequencing, the 1st 8 bases of read identifies which sample it came from. Some Introductions: What do you want to learn from this course? What do you already know about RNA-Seq? DAY 1 LECTURE OUTLINE • ChIP method, validating your antibody. • Next Generation Sequencing technology • Preparing a ChIP-seq library • Choosing options for sequencing • Looking at published ChIP-seq results -Exercise: Tracks on the UCSC browser Preparing ChIP-seq Libraries Step 1: Quantitate your ChIP recovery Typical ChIP recoveries are very low: … on the order of 2 to 10 ng of total recovered DNA from 100 microliters of tissue or packed cells (about what you’d get from a 10 cm tissue culture plate at confluence). Need a method to quantitate your recovery that is sensitive down to ~0.1 ng/microliter. UV, ethidium bromide & pico green are not good enough. I use InVitrogen’s Quant-IT dsDNA HS Assay Kit. Requires a flourescence microplate reader. If your recovery is >3ng you should be able to make a library easily enough. If it is <2 ng you can try to make the library & if it passes QC it might be fine, or you may want to scale up your ChIP. Preparing ChIP-seq Libraries Steps 2 through 4: End repair & Adapter Ligation Sonicated fragments (from ChIP & equal weight of input control DNA) End repair (3’ overhang exonucleases + 5’ overhang fill-in) A-tailing (Taq polymerase adds terminal A) For ChIP2 or ChIP-seq, it is preferable to use purified input chromatin fragments for control library construction. The IgG negative is less useful for ChIP-seq (but is important to use in initial ChIP experiments to establish the effectiveness & specificity of your antibodies!). Adapter ligation (Illumina adapters & T4 DNA ligase) Ethanol precipitate with ultrapure glycogen carrier, to reduce volume & get into buffer that won’t interfere with agarose gel run Preparing ChIP-seq Libraries Step 5: First Size Selection (removing unligated adapters) Electrophoresis on agarose gel works fine. You won’t see a band (you started with only 3 ng of DNA!), so you’ll have to choose a range to cut with reference to MW markers. [you may see some signal for the adapters running at ~90 bp] •Flexible, easy to do w/ standard lab supplies •Prone to contamination: so run samples with spacer lane between them. •Qiagen gel extraction kit recovery is usually acceptable Other methods of size selection: Invitrogen E-gel System (agarose separation w/ collection window) •Can be faster than normal agarose gel but… •Collection in narrow size range (~10-20bp) & hard to collect in larger range •Recoveries not much better than agarose Closed cell systems w/ dynamic collection Pippin Prep Caliper LabChip XT Great if you can afford them (very expensive!) …fortunately, in my experience, Pippin Prep versus Qiagen gel extraction kit showed comparable recoveries. Step 6: Limited Amplification •A tail establishes orientation for primer. 5’ 3’ 5’ 3’ •1st: primer matching p7 creates complete duplex. •Next: P7 & P5 primers amplify it. Optimize your PCR cycles Goal is to minimize PCR bias (some fragments amplifying more than others) Perform 9 cycles of PCR remove aliquot Perform 3 cycles of PCR remove aliquot … Repeat for 18 total cycles ->Agarose gel Find cycle number that gives strong product with few primer dimers, do multiple reactions w/ that cycle number & pool. EtOH ppt. w/ glycogen carrier. Specific product (should be fragsize+~90bp for adapters) Primer dimers, etc. (<200bp, often <100) Step 7: Final gel purification Isolate band & purify (~20-50 bp range best, but can take larger if you need more recovery) Quantitate recovery (can use DNA HS flourescence kit, but, ideally you’ll have enough recovery to measure even by spectrophotometer). Need ~10 microliters of 10 ng/microliter sample for sequencing (a bit less might be OK… contact your core for their requirements & opinions). Gel should be new w/ clean buffer & lanes separating samples. Biological Repeats •NGS technology is very robust & less prone to the day to day or array to array variability that plagues microarrays. •Your major sources of variability will be: 1) -Biological variability (best to have 3, or at least 2, biological replicates). 2) -Library preparation (need to be very careful to prepare libraries the same way, ideally on the same day). Minimizing technical variation & contamination •Order all reagents needed for the experiment to ensure consistency (don’t use old lab stocks) •Make all solutions fresh (from 1st fixation step before ChIP onwards) •Use low retention, filter tip pipettes at every step •Ideally, perform the library prep for all samples simultaneously •Follow exact same protocol, especially including size ranges isolated at each gel purification. DAY 1 LECTURE OUTLINE • ChIP method, validating your antibody. • Next Generation Sequencing technology • Preparing a ChIP-seq library • Choosing options for sequencing • Looking at published ChIP-seq results -Exercise: Tracks on the UCSC browser How many reads do I need? Minimum for ChIP-seq of a transcription factor with < ~30,000 binding sites in a mammalian genome: • 2 replicates per condition • 20+ million reads per sample (>40M per condition, proportionately less for smaller genomes & fewer binding peaks) • One HiSeq lane gives ~150 million reads …& can multiplex ~4 samples (2 exp + 2 input / lane) • Single end 50 bp reads almost always good (unlike RNA-seq where longer and/or paired end reads are required for many downstream questions). For some applications need many more reads (e.g. mapping nucleosome positions need >400 M). Make your best estimate. If you have too few you can re-sequence the same samples or add additional samples. Reads from all runs can be pooled in the end. How much will it cost? Multiplexing 4 samples per lane Library prep costs ($~200 per sample) Current Tufts Genomics Core Probably around $2000 total per 2 biological replicates of one condition. Can save by using one input sample as background for several replicates or conditions, but this assumes fragmentation was virtually identical across all samples (not recommended unless you’re really confident in your technique). The TUCF Genomics Core: http://genomics.med.tufts.edu Sign up for an account. Login & click “create new order” For questions about sample preparation and Illumina protocols, please contact tucf-ngsprep@tufts.edu. For all other questions about the service, including scheduling and consulting, please contact: Albert Tai Genomics Core Manager Tufts University School of Medicine 150 Harrison Ave, Jaharis 523A Boston, MA 02111 617-636-3992 albert.tai@tufts.edu DAY 1 LECTURE OUTLINE • ChIP method, validating your antibody. • Next Generation Sequencing technology • Preparing a ChIP-seq library • Choosing options for sequencing • Looking at published ChIP-seq results -Exercise: Tracks on the UCSC browser ChIP-seq for histone modifications Method: •Mouse ES cells vs ES-derived primary neural progenitor cells (NPCs). •Prepare chromatin & ChIP with antibodies to specific histone modifications. •H3K4 methylation marks active genes, H3K27 marks repressed genes, both marks together in ES cells mark “poised” genes that will become activated in certain developmental lineages. Meissner et al. 2008, Nature 454:766. The ENCODE Project Dozens of labs did ChIP-seq, under rigorous quality guidelines, for over 100 transcription factors and histone modifications, plus related assays for DNA methylation, chromatin accessibility etc. Major paper (many others provide additional details): Encode Project Consortium (over 100 authors) An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):5774. doi: 10.1038/nature11247. Some ways to access this data: Nature.com/encode (Nature’s summary & links to all related papers) factorbook.org (a way to explore the data in a wiki format) UCSC genome browser (hands on examples next) sra (short read archive, repository for raw data, more on this later!) Sample of Encode Data Accessing Encode ChIP-seq data Start up your browser: Go to: http://genome.ucsc.edu Click on “genomes” tab on top left Select Human and hg19 Hit [submit] button Look around & familiarize yourself with the controls: Click & drag on line with bp numbers-> zoom selection, 1x, 3x, 10x zoom options, control click on track gives options: keep refseq genes track & “hide” others Scroll down to blue bar that says “regulation” Click ENC histone (for Encode histone modification tracks) box to “show” Then click on “Enc histone” blue underlined name-> opens controls. Check “Broad histone” and set to “full”, & click on its name -> opens specific controls. Columns are ChIP-seq with different antibodies, rows are different cell lines. Check “peaks” dense & “signal” full. Uncheck any pre-checked boxes & then check, H3K4me3, H3K27ac & H3K27me3 in cell lines H1-hESC (embyronic stem cells), HepG2 (liver) & Osteoblasts. Then go back to top & hit [submit] At box on top type in “mtrf1l” & hit return, choose the top match. Click zoom out 3x, & then zoom out 10x (on top right) Example of ChIP-seq data tracks on UCSC browser Peak calls: regions of significant enrichment over background Processed read density, read as # of reads overlapping a given BP position, data (used to make peak calls) H3K4me3 & H3K27acet are marks of active promoters (e.g. MTRF1L) H3K27me2 is a mark of repressed promoters Genes in ESCs that are required for differentiation are often “poised” and bear K4me3 & K27me3 “bivalent mark” (e.g. SYNE1) Differentiation resolves bivalent mark to all activating marks (Osteoblasts) or all repressive marks (HepG2) Downloading data from UCSC browser Try zooming in (you can go all the way to base pair resolution) Want to learn more about a gene? Control click on it’s ideogram & select “open details page in new window” What if you want to use this data somewhere else? Select Tools->Table Browser Select Group: Regulation, Track: Broad Histone Table: H1-hESC H3K4me3 … Pk (for the peaks data, the signal file will be huge) In output format, select “all fields from selected table” --Note that you could have selected “sequence”… if you had you’d get the actual DNA sequence for each one of these peaks. We’ll use this later. Check “Galaxy” next to send output to: --Note that you could have selected send to file, we’ll use this later as well. Click “Get output” & then click “send query to galaxy” Introduction to Galaxy Tools Galaxy is a web platform providing a lot of basic tools for manipulating genomics data. On the right are input & analysis options. On the left is your history of uploaded files & analyses. You’ll have one item in process, which will finish soon & turn green. Click on the title to get a sample of what the data looks like. Click on the eye to see the data in the central panel. Each entry has a chromosome#, BP for start & BP for end & some other values (signal value=enrichment over background, p.value=log(base10)of p. value, so, for p=.0001 this would be 4) Click on the pencil to look at and edit the name & other attributes of any item. We’ll look more at Galaxy tools later… What about data that’s not on the UCSC Browser? The ENCODE project was UNUSUALLY considerate when compared to most other researchers who generate genomics data. Even though ENCODE is huge, it’s probably <10% of published NGS data. To publish, researchers must make their data accessible, but they will very rarely provide a link to a UCSC browser track. If you’re lucky, they will have put processed data up somewhere: generally on GEO… The GENE EXPRESSION OMNIBUS (GEO) Key repository for microarray and genomics data. Open a new browser tab (ctrl-T) & go to: http://www.ncbi.nlm.nih.gov/geo/ Search for “encode h3k4me3 h1-hesc”. You’ll see several entries. The first few are larger datasets that include this specific data. The one at the bottom is just the data for this track. First note the “accession number GSM733657” - often publications will give this accession number, providing an easy way to get directly to the right place. ---> Now, click on the title for this entry “Bernstein…” Scroll down the next page: There’s lots of info about this experiment with links for more information. At the bottom are the processed data files: They’ve been nice & offer us a “BROADPEAK” file (the same as what we just uploaded to Galaxy), a “BAM” file for each experimental replicate (which has the genomic coordinates for each read), and a “BIGWIG” file (the filetype for the “signal” track on the UCSC browser) What if the data I’m interested in isn’t in GEO? Authors are almost always required to make their NGS data accessible in order to publish…. …but they’re often not required to make it easy! Many times the only thing that’s available is the raw data, stored in… The Sequence Read (SRA) - Repository for data. Open a new browser tab (ctrl-T) & go to: raw NGS http://www.ncbi.nlm.nih.gov/sra/ Search again for “encode h3k4me3 h1-hesc”. A single record will be called up… Clicking on link 1 or 2 under “run” will give you information about that particular biological replicate sample. Clicking on the link 1 or 2 under ‘size’, will take you to a page with a single linked file with a “.sra” extension. Note how big this file is… just a few of these would rapidly fill up a PC hard drive! So what is a .sra file & what can you do with it? Don’t even try to download & open it… besides being huge, it’s not even normal text. In the next few lectures we’ll find out how to handle this sort of raw data. SEQanswers Answers to your questions about NGS applications Forums (ask Q’s &/or search for A’s) Wiki – Find NGS software Instrument Map ChIP-seq COURSE OUTLINE • Day 1: ChIP techniques, library production, USCS browser tracks • Day 2: QC on reads, Mapping binding site peaks, examining read density maps. • Day 3: Analyzing peaks in relation to genomic feature, etc. • Day 4: Analyzing peaks for transcription factor binding site consensus sequences. • Day 5: Variants & advanced approaches. ChIP-seq for TF (SISSRS software) Jothi, et al. Genome-wide identification of in vivo protein– DNA binding sites from ChIP-Seq data. NAR (2008), 36: 5221-31 Adapted from slide set by: Stuart M. Brown, Ph.D., Center for Health Informatics & Bioinformatics, NYU School of Medicine