ChIP-seq Methods & Analysis
advertisement

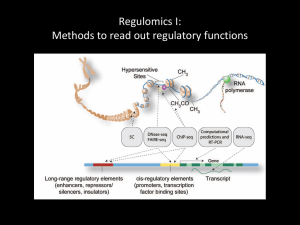
ChIP-seq Methods & Analysis Gavin Schnitzler Asst. Prof. Medicine TUSM, Investigator at MCRI, TMC gschnitzler@tuftsmedicalcenter.org 617-636-0615 ChIP-seq COURSE OUTLINE • Day 1: ChIP techniques, library production, USCS browser tracks • Day 2: QC on reads, Mapping binding site peaks, examining read density maps. • Day 3: Analyzing peaks in relation to genomic feature, etc. • Day 4: Analyzing peaks for transcription factor binding site consensus sequences. • Day 5: Variants & advanced approaches. Day 5 Outline • Introduction to variations on ChIP-seq methods • Extensions & variations on TFBS analysis • Analyzing published data & across platforms • Downloading & installing programs • Writing your own programs Next-Generation Sequencing Analysis “ChIP-Seq is the best thing that happened to ChIP since the antibody. It is 100x better than ChIP-Chip since it escapes most of the problems of microarray probe hybridization. Plus it is cheaper, and genome wide. But ChIP-Seq is only the tip of the iceberg - there are many inventive ways to use a sequencer.” Quote from intro to Homer software at: http://biowhat.ucsd.edu/homer/ngs/index.html Extensions of ChIP-seq ChIP-Seq: Isolation and sequencing of genomic DNA "bound" by a specific transcription factor, covalently modified histone, or other nuclear protein. This methodology provides genome-wide maps of factor binding. Most of HOMER's routines cater to the analysis of ChIP-Seq data. DNase-Seq: Treatment of nuclei with a restriction enzyme such as DNase I will result in cleavage of DNA at accessible regions. Isolation of these regions and their detection by sequencing allows the creation of DNase hypersensitivity maps, providing information about which regulatory elements are accessible in the genome. (variant technique called FAIRE-seq) MNase-Seq: Micrococcal Nuclease (MNase) is a restriction enzyme that degrades genomic DNA not wrapped around histones. The remaining DNA represents nucleosomal DNA, and can be sequencing to reveal nucleosome positions along the genome. This method can also be combined with ChIP to map nucleosomes that contain specific histone modifications. RNA-Seq: Extraction, fragmentation, and sequencing of RNA populations within a sample. The replacement for gene expression measurements by microarray. There are many variants on this, such as Ribo-Seq (isolation of ribosomes translating RNA), small RNA-Seq (to identify miRNAs), etc. GRO-Seq: RNA-Seq of nascent RNA. Transcription is halted, nuclei are isolated, labeled nucleotides are added back, and transcription briefly restarted resulting in labeled RNA molecules. These newly created, nascent RNAs are isolated and sequenced to reveal "rates of transcription" as opposed to the total number of stable transcripts measured by normal RNA-seq. Hi-C: Genomic interaction assay for understanding genome 3D structure. This assay is much more specialized - For more information about how to use HOMER to analyze Hi-C data, check out the Hi-C analysis section. Examining longrange interactions by ChIP-seq Two DNA fragments associated with the same IP’d protein are ligated together. Sequencing identifies both short-range and long range interactions. Nature Reviews Genetics 2012 13:840 Fine scale information from DNAse-seq Sequencing the ends of DNAse cuts identifies regions of bare DNA. Fine scale analysis of this data can identify individual TF binding sites. Nature Reviews Genetics 2012 13:840 Capturing allele-specific information using SNPs in reads CTCF binds better to the A variant Mapping CpG DNA methylation patterns Approaches: •IP of DNA fragments using antibodies against meC or meCpG binding proteins. •Selection of DNA fragments using methyl-sensitive restriction enzymes. •Whole genome bisulfite sequencing. Bormann Chung CA, Boyd VL, McKernan KJ, Fu Y, et al. (2010) Whole Methylome Analysis by Ultra-Deep Sequencing Using Two-Base Encoding. PLoS ONE 5(2): e9320. doi:10.1371/journal.pone.0009320 http://www.plosone.org/article/info:doi/10.1371/journal.pone.0009320 Mapping nucleosome positions Approaches: •1) Fragmentation to mononucleosome size by sonication or micrococcal nuclease (MNase) ChIP w/ antibody against histone modification (H3K4me1) – can map positions of nucleosomes with this mark. Whole genome sequencing. Nat Struct Mol Biol. 2011 June; 18(6): 742–746. Plotting ChIP-seq read density versus genomic features Taking average normalized .bedgraph data relative to TSSes… ChIP-seq reads/10bp/promoter 60 LiERBS_v_LiD LiERBS_v_LiU LiERBS_v_LiNon-regl. LiERBS_v_LiNon-expr. AoERBS_v_AoD AoERBS_v_AoU AoERBS_v_AoNon-regl. 50 40 30 20 10 0 -2000 -1000 0 1000 BP from TSSes of gene group 2000 Using input chromatin read density to measure nucleosome densities Hypothesis: Sonication mostly cuts in nucleosome free regions or internucleosomal spacers. Thus, read positions give information about nucleosome positions. Initial support: Average normalized .bedgraph data from INPUT sample relative to TSSes recapitulates the low nucleosome occupancy seen genomewide over promoters. Li INPUT shifted reads/ 10 bp/ promoter 8 7 6 5 LiINPUT_v_LiD_pros (norm'd) LiINPUT_v_LiU_pros (norm'd) LiINPUT_v_AoD_pros (norm'd) LiINPUT_v_AoU_pros (norm'd) 4 3 2 1 0 -2000 -1500 -1000 -500 0 500 BP from Li Down promoter TSSes 1000 1500 2000 Day 5 Outline • Introduction to variations on ChIP-seq methods • Extensions & variations on TFBS analysis • Analyzing published data & across platforms • Downloading & installing programs • Writing your own programs Many approaches to TFBS analysis Hannenhalli S Bioinformatics 2008;24:1325-1331 Also, Ladunga I. An overview of the computational analyses and discovery of transcription factor binding sites. Methods Mol Biol. 2010;674:1-22. doi: 10.1007/978-1-60761-854-6_1. : Introduction to a set of about a dozen methods papers. De Novo Search Algorithms The Gibbs sampler approach 1 k Objective: Find conserved segment of length k in n unrelated 1 k sequences The program will need to run once for each k: e.g. 6 bp, 7 bp, 8 bp sequences, etc. (either automatically, or by hand). 1 1 k From : Lawrence, C. et al.(1993) Detecting Subtle Sequence Signals: A Geibbs Sampler approach to Multiple Alignment. Science 262.208- The EM approach (in MEME etc.) Expectation Maximization algorithm, proceeds in iterations until E & M converge. For an explanation of the process see Nature Biotechnology 26, 897 - 899 (2008). Adapted from: www.stats.ox.ac.uk/~hein/Signals.ppt 2 n Two de novo search methods • DME is part of the same CREAD package that storm is in (run in UNIX) • SEME some of the same refinements as CentDist to do de novo searches: http://biogpu.d1.comp.nus.edu.sg/~chip seq/webseqtools2/ Extensions to Basic Models Composite Patterns: BioOptimizer: the Bayesian Scoring Function Approach to Motif Discovery Bioinformatics M2 Start p12 Regulatory Modules: p21 De novo cis-regulatory module elicitation for eukaryotic genomes. Proc Nat’l Acad Sci USA, 102, 7079-84 M3 M1 Gene A Gene B Stop Adapted from: www.stats.ox.ac.uk/~hein/Signals.ppt Combining Signals and other Data Motifs Coding regions Expresssion and Motif Regression: Integrating Motif Discovery and Expression Analysis Proc.Natl.Acad.Sci. 100.3339-44 1.Rank genes by E=log2(expression fold change) 2.Find “many” (hundreds) candidate motifs 3.For each motif pattern m, compute the vector Sm of matching scores for genes with the pattern 4.Regress E on Sm Yg m Smg g ChIP-on-chip - 1-2 kb information on protein/DNA interaction: An Algorithm for Finding Protein-DNA Interaction Sites with Applications to Chromatin Immunoprecipitation Microarray Experiments Nature Biotechnology, 20, 835-39 Protein binding in neighborhood Coding regions Adapted from: www.stats.ox.ac.uk/~hein/Signals.ppt Assessment of evolutionary conservation Modules shared across species are most highly rated. For use of evolutionary conservation information w/ individual motifs see: Das & Dai 2007 BMC Bioinformatics 8:S21. For regulatory modules see: Su J, Teichmann SA, Down TA (2010) Assessing Computational Methods of Cis-Regulatory Module Prediction. PLoS Comput Biol 6(12): e1001020. doi:10.1371/journal.pcbi.1001020 http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1001020 Adapted from: www.stats.ox.ac.uk/~hein/Signals.ppt Integrating data from multiple sources w/ permutation of average ranks Let’s say we want to combine data from several sources or metrics to decide which are the most relevant enriched TFs. e.g. 1) p.value in CentDist, 2) p.value in Storm & 3) p.value of homologous sequence in DME Establish a ranking metric for each (e.g. 1 best to 10 worst). It doesn’t have to be the same for 1, 2 & 3, but you need to apply the same rank system across different biological conditions. For each TF compute the average rank. (1) 1 4 3 8 . (2) 3 5 4 9 . (3) 2 8 2 7 . (avg) 2 5.7 3 8 . Permutation of average ranks Now take the same columns of ranks for (1), (2) & (3) and randomize each one separately. Repeat this several times (until you have thousands of random average ranks & plot frequency vs avg. rank… (1) 8 4 3 1 . (2) 5 4 3 9 . (3) 2 8 7 2 . (avg) 5 5.3 4.3 4 . 2.0 observed 34/10,000 times in permuted averages. Estimated FDR ~3.4e-3 10 7 5 3 1 The number of times a given value is observed divided by the total number of iterations gives an estimate of false discovery rate. Day 5 Outline • Introduction to variations on ChIP-seq methods • Extensions & variations on TFBS analysis • Analyzing published data & across platforms • Downloading & installing programs • Writing your own programs What if you want to know something from a published dataset, but they’ve only provided the raw data on SRA? Getting data from SRA Go to: http://www.ncbi.nlm.nih.gov/sra Find an experiment by searching, e.g. “encode h1-hesc h3k4me3” Click on the name to the left of the smaller file (1.9M) & then on the downloads tab. Right click on the ftp link for the run & copy the link location. Open putty & login to your account at cluster.uit.tufts.edu Go to your /cluster/shared/[userID]/chip directory & do: wget [pasted URL] Decoding the .sra format The SRR227387.sra file you now have is in a special file format, but it does have all the original .fastq information in it. To get that info do: bsub /cluster/tufts/cbi*/Ch*/ESC*/sra*/bin/fastq-dump SRR227387.sra [fastq-dump is part of a package of programs for handling .sra files that you can download, unpack & run immediately from your shared directory – at least as far as simple files like fastq-dump are concerned] This gives you the same .fastq format you’re familiar with. Use head to confirm the format, but then you might as well delete the file with rm so as not to clutter up the cluster. After this week you are now ready to do any analysis you want on this data, from mapping reads to the genome (w/ bowtie) to peak calling (w/ MACS), to TFBS analysis. “Liftover” programs to convert between genomes & builds Several useful tools for this in Cistrome/Galaxy: Liftover/Others Convert between RefSeq, Gene Symbols to Entrez IDs using Bioconductor. Liftover Wig Files Liftover wig files [Galaxy]Convert genome coordinates between assemblies and genomes Extract data from Wiggle Extract data for certain chromosome from a wiggle file Extract data from Bed Extract data for certain chromosome from a BED file In the UCSC genome browser: •Tools-> Liftover •Choose the starting genome/build & the one you want to convert to. •Upload a .bed file w/ the ranges you want & hit go (only works for bed files… may work with bedGraph, although I haven’t confirmed this) Day 5 Outline • Introduction to variations on ChIP-seq methods • Extensions & variations on TFBS analysis • Analyzing published data & across platforms • Downloading & installing programs • Writing your own programs Don’t be intimidated! There’s nothing to prevent you from installing a program you want to run in your cluster account. Before you begin, though, type “module available” to see if it’s already installed as a module. Also go to /cluster/tufts/ngsp/ngsp/ to see if it’s installed there. • Read the documentation from the creator’s lab, download, unzip &/or unpack the file, read the INSTALL or README files included, & give it a try. • You may need to be running a specific version of perl or python, etc. If so, check “module available” to see if it’s installed on the cluster & use “module load [name]” to add it. • You may also need to set system variables using “export VARIABLE=$VARIABLE:/new/path”. README files should tell you enough to know what to try. • If you get stuck, the cluster support folks are friendly & helpful (and respond moderately fast). Contact them at: cluster-support@tufts.edu. A different integrated package of tools to run in UNIX HOMER Software for motif discovery and next-gen sequencing analysis http://biowhat.ucsd.edu/homer/ngs/index.html Mapping to the genome (NOT performed by HOMER, but important to understand) Creation Tag directories, quality control, and normalization. (makeTagDirectory) UCSC visualization (makeUCSCfile, makeBigWig.pl) Peak finding / Transcript detection / Feature identification (findPeaks) Motif analysis (findMotifsGenome.pl) Annotation of Peaks (annotatePeaks.pl) Quantification of Data at Peaks/Regions in the Genome/Histograms and Heatmaps (annotatePeaks.pl) Quantification of Transcripts (analyzeRNA.pl) Additional analysis strategies: General sequence manipulation tools (homerTools) Miscellaneous Tools for Sharing Data between programs, etc. (tagDir2bed.pl, bed2pos.pl, pos2bed.pl ...) Finding overlapping or differentially bound peaks (mergePeaks, getDifferentialPeaks) ChIP-Seq analysis automation (analyzeChIP-Seq.pl) Description of file formats Could be very useful… & with (only a bit of) luck, you’ll be able to install & run them yourself. Installing a program in R Check out the Key R Commands link at http://sites.tufts.edu/cbi/resources/chip-seq/ This is not an introduction to programming in R! Instead it gives basic instructions for how to: 1) install & run R packages that may be needed for your research, 2) how to move data files into R 3) how to perform simple edits on this data that may be required by the package & 4) how to output your results. Note: I find that the documentation for R packages is generally quite good. Day 5 Outline • Introduction to variations on ChIP-seq methods • Extensions & variations on TFBS analysis • Analyzing published data & across platforms • Downloading & installing programs • Writing your own programs Mastering simple UNIX tools find, awk, grep, sort, sed & more One line commands to let you search and manipulate large data files w/o writing a program or trying to use the kludgy and limited tools in Galaxy. Find out more at: http://sites.tufts.edu/cbi/resources/rna-seqcourse/unix-resources/ Programming: Get your feet wet Perl Tutorials - learn.perl.org learn.perl.org/tutorials/ Many tutorials are available if you are interested in learning Perl. These tutorials are introductions. Beginning Perl (free) - www.perl.org www.perl.org/books/beginning-perl/ This book is for those new to programming who want to learn with Perl. A ton of Perl programs for you to use/adapt/modify: http://www.bioperl.org/wiki/Main_Page For learning R: Check out Josh’s links at: http://sites.tufts.edu/cbi/resources/rnaseq-course/r-resources/ Also check out my notes on using R (specifically geared to the minimum you need to install & use existing programs) & a brief reference sheet on Perl at http://sites.tufts.edu/cbi/resources/chip-seq/ Look at examples, check the web… If you’re looking for a command in UNIX, R, Perl, Python, etc. do a Google search (for R add “statistical” to your search to specify what you mean). If you’re wondering how to get a program to do something, look at other programs & see how they did it. You don’t need to memorize the language, beyond a few basics, just look at what you (or someone else) did before & copy it. Questions? What would you like to explore? What’s the next bioinformatics challenge in your research? Course evaluation forms…