Big Idea 3 – Investigation (Lab)
advertisement

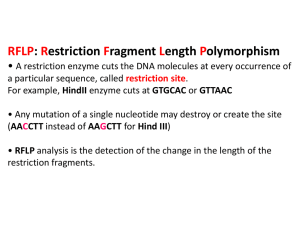
Big Idea 3 – Investigation (Lab) 9 We did a variation of this lab at the Cold Spring Harbor DNA Learning Center West… The next few slides highlight the differences between what you did and what this lab specifically asked that you do…which is to pretend that you are working a crime scene, have a DNA sample, and will cut with restriction enzymes to look for restriction fragment length polymorphisms (RFLP’S) Big Idea 3 – Investigation (Lab) 9 Crime scene DNA Suspect DNA Restriction fragment length polymorphisms (RFLP’s = “rif lips”) The differences in restriction sites found on homologous chromosomes giving rise to different numbers and lengths of restriction fragments...review chapter 20 powerpoint for more detail. Amplified section of the Amplified section of the DNA from the crime scene same DNA segment from the Big Idea 3 – Investigation (Lab) 9 IDEAL GEL A gel showing lambda phage DNA (same DNA you cut at the DNA learning center) cut with three different restrictions enzymes and an uncut negative control. Look at chapter 20 for a review of how gel electrophoresis works… Big Idea 3 – Investigation (Lab) 9 Calculating the Standard Curve Big Idea 3 – Investigation (Lab) 9 Calculating the Standard Curve 2.3 2.3 3.0 3.5 4.2 5.8 6.3 Big Idea 3 – Investigation (Lab) 9 Calculating the Standard Curve Size of fragment (bp) Relationship between DNA fragment size in bp and migration distance in a 0.8% agarose gel. 25000 2.3 2.3 3.0 3.5 4.2 5.8 6.3 20000 15000 10000 5000 0 0 2 4 6 8 Migration distance (cm) There appears to be an exponential relationship… Or…small fragments move exponentially quicker than larger fragments… If so, then using a log scale on the y-axis should result in a linear relationship…(next slide) Big Idea 3 – Investigation (Lab) 9 Calculating the Standard Curve 100000 10000 1000 100 2.3 2.3 3.0 3.5 4.2 5.8 6.3 10 1 0 2 4 6 8 20000 R2=0.93057 2000 0 2 4 6 8 Big Idea 3 – Investigation (Lab) 9 Calculating the Standard Curve 20000 R2=0.93057 2.3 2.3 3.0 3.5 4.2 5.8 6.3 2000 0 2 4 6 8 This is now our standard curve…we can use it to do what? To calculate the sizes of the BamHI and EcoRI bands… Simply measure the distance traveled by the band and then use the curve to determine the size. Big Idea 3 – Investigation (Lab) 9 Calculating Size of Unknown Bands 20000 14,000 2.3 2.3 3.0 3.5 4.2 5.8 6.3 2.5 R2=0.93057 14,000 2000 0 2 4 6 8 Calculate the size of the largest fragment for the BamHI digest. The migration distance is 2.5 cm. Therefore, the size is approximately 14,000 bp (I actually used the formula for the line, which excel can give you, instead of the graph itself, but the AP will require you to use the graph). Big Idea 3 – Investigation (Lab) 9