Analysis of genome-wide
association studies
Lecture 1: Introduction
Linkage studies
• Traditional approach to identifying genes for human
traits and diseases was through linkage.
• For Mendelian diseases (e.g. Huntington’s disease)
there is a clear co-segregation of genetic markers
with disease within pedigrees.
• For complex traits (e.g. type 2 diabetes), linkage
analysis has been less successful because the
relationship between phenotype and genotype is
less clear.
• Non-genetic risk factors influence the outcome;
• Many genes have an impact on the trait, each having only
a small effect on the outcome.
Association studies
• Common disease, common variant hypothesis:
complex traits will be determined by variants that
occur frequently in the population, but each have only
a small impact.
• Ascertain sample of affected cases and unaffected
controls (or a random sample for a quantitative trait)
from the population.
• Compare allele frequencies in cases and controls (or
mean trait values between alleles for a quantitative
traits).
• With sufficient sample size, powerful approach to
identify loci contributing to complex traits.
Common variation in the genome
• Impractical to genotype all common variants
in the genome in large samples.
• International HapMap project genotyped
more than three million SNPs in samples from
multiple ancestry groups.
• Common variation is arranged on relatively
few haplotypes that occur within blocks of
strong linkage disequilibrium between
recombination hotspots...
Common variation in the genome
BLOCK 1
SNPs in strong LD
Low diversity of
haplotypes
BLOCK 3
HOTSPOT
Low diversity of
haplotypes
HOTSPOT
SNPs in strong
LD
BLOCK 2
SNPs in strong LD
Low diversity of
haplotypes
• SNPs located within the same block will be in strong LD with each
other. However, a pair of SNPs located in adjacent blocks will be
uncorrelated due to the high levels of recombination at the flanking
hotspot.
• The strong LD between SNPs in the same block means that we tend
to observe fewer haplotypes than we might expect by chance. In
fact, much of the diversity is accounted for by a small number of
common haplotypes.
Common variation in the genome
• Jeffreys et al. (2001) looked for evidence of recombination in the sperm of
six men across ~200kb region of the MHC.
• Crossovers cluster in six hotspots: recombination extremely rare elsewhere
in the region.
• High-resolution LD analysis in sample of 50 unrelated individuals identified
blocks with boundaries corresponding to these recombination hotspots.
(Nature Genetics 29: 217-222)
3-
Common variation in the genome
• Dawson et al. (2002) genotyped 90 unrelated UK individuals at over 1500 SNPs
across chromosome 22.
• Defined haplotype blocks as regions of limited haplotype diversity.
(Thanks to Lon Cardon)
Common variation in the genome
Genotyping this subset of SNPs accounts for all common
genetic variation in the block
Measures of LD
• Linkage disequilibrium between two marker allele M and
disease variant X quantified by measure:
D P MX
P M P X
• Adjustment for allele frequencies:
D
D
D MAX
r
2
D
2
P M P m P X P x
• May not have typed disease variant: but understanding
patterns of LD between markers will help interpretation of
results of association studies.
Genome-wide association arrays
• Possible to “cover” much of the common
genetic variation in the human genome by
genotyping only a subset of SNPs.
• Most efficient approach is to select “tags” that
cover common SNPs (at some r2 threshold).
11
• Samples genotyped with
Affymetrix GeneChip 500K
Mapping Array Set.
• Identified novel loci and
replicated association signals
for five diseases.
• Established standard protocols
for quality control and analysis
of GWAS.
• Publicly available control
cohort.
Published Genome-Wide Associations through 07/2012
Published GWA at p≤5X10-8 for 18 trait categories
NHGRI GWA Catalog
www.genome.gov/GWAStudies
www.ebi.ac.uk/fgpt/gwas/
Design issues
• Is the trait heritable?!
• Phenotype definition, case-control selection,
and availability of non-genetic risk factors.
• Choice of genotyping product.
• Sample size requirements.
• The GENETIC POWER CALCULATOR
(http://statgen.iop.kcl.ac.uk/gpc) can be used
to calculate power of case-control studies.
QUALITY CONTROL
Introduction
• Poor study design and errors in genotype
calling can introduce systematic bias in
association studies.
• Increase in false positive error rate and decrease
in power.
• Assess data quality to remove sub-standard
genotypes, samples and SNPs from
subsequent association analysis.
Genotype calling
• For large-scale GWA studies, automated genotype
calling algorithms have been developed, e.g.
GENCALL and GENOSNP.
• Estimate probability that any specific genotype is AA, AB or
BB.
• Apply threshold to probabilities in order to call genotype,
otherwise treated as missing.
• Choice of calling threshold will impact results:
• Too low: include poor quality genotypes.
• Too high: unnecessarily remove high quality genotypes, or
may introduce bias by preferentially calling specific
genotypes (e.g. rare homozygotes).
Genotype calling
Genotype calling
• For large-scale GWA studies, automated genotype
calling algorithms have been developed, e.g.
GENCALL and GENOSNP.
• Estimate probability that any specific genotype is AA, AB or
BB.
• Apply threshold to probabilities in order to call genotype,
otherwise treated as missing.
• Choice of calling threshold will impact results:
• Too low: include poor quality genotypes.
• Too high: unnecessarily remove high quality genotypes, or
may introduce bias by preferentially calling specific
genotypes (e.g. rare homozygotes).
Sample quality control
• Remove samples on the basis of:
• Low call rate (poor DNA quality).
• Outlying heterozygosity across autosomes (DNA
sample contamination or inbreeding).
• Duplication or relatedness based on identity-bystate (samples should be independent).
• Mismatches with external information (sample
mix-up).
• Outlying population ancestry (confounding due to
population structure).
Call rate and heterozygosity
23-30% heterozygosity
3% missing
Identity-by-state (IBS)
• Over M markers, the IBS between the ith and jth individuals is
given by
IBS ij 1
1
2M
G
ik
G jk
k
where Gik denotes the number of minor alleles (0, 1 or 2)
carried by the ith individual at SNP k.
• Identical samples will share IBS near to 100% (allowing for
genotyping errors).
• Related individuals will share higher IBS than unrelated
individuals.
• Common to plot histogram of IBS of each individual with
“nearest neighbour”.
IBS distribution
Remove one sample from each duplicate or related
pair (usually one with lowest call rate).
Duplicates
Relateds
X chromosome
• Distribution of heterozygosity different in
males and females.
• Should be no heterozygosity in males, but expect
some genotyping error.
• Discrepancies with external gender
information may reflect:
• Errors in external data;
• Sample mix-up;
• Gender inconsistent with sex chromosomes.
X chromosome
Each individual plotted twice
according to reported gender:
females in red and males in blue.
Should these samples be removed
from the study or the sex
corrected based on
heterozygosity? May impact on
results if sex is adjusted for in the
analysis or if sex specific analyses
are to be undertaken.
SNP quality control
• Remove SNPs on the basis of:
• Low call rate, variable by MAF (poor quality SNP).
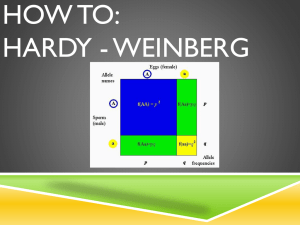
• Extreme deviation from Hardy-Weinberg equilibrium in
cases, controls or both for autosomes (genotyping error).
• Extreme differential call rates between cases and controls
(calling bias).
• Study specific SNP QC filters (such as differences in allele
frequencies between multiple control cohorts).
• Low frequency SNPs (more prone to bias due to
genotyping error and low power to detect association).
• Visual inspection of cluster plots.
Effect of differential call rate
Individuals called as missing?
Fewer heterozygotes among cases.
Cluster plot inspection: good SNP
Cluster plot inspection: bad SNP
Summary
• QC criteria are subjective and vary from one study to
another.
• Sample QC filters should not be so stringent as to
remove the majority of the analysis cohort!
• SNP QC filters should eliminate the worst quality
markers without “throwing the baby out with the
bathwater”.
• All SNPs demonstrating evidence for association
should be followed up with visual inspection of
cluster plots.
BASIC ANALYSIS OF GENOME-WIDE
ASSOCIATION STUDIES
Introduction
• Association analyses focus on the
identification of SNPs that differ in allele
(genotype) frequency between cases and
controls.
• Basic analyses utilise standard epidemiological
tools, rather than specialised methods that
have been developed for analysing more
traditional pedigree and family studies:
• contingency table analysis;
• logistic regression modelling.
Genotype-based test
•
•
Assuming the sample to be typed at a SNP
marker of interest, we can represent
genotype data in a 2 x 3 contingency table.
2
The usual test for independence of rows
and columns in contingency tables can be
applied to test the null hypothesis of no
disease-marker association
X
2
i 0 ,1 , 2 j A ,U
where
•
E n ij
n
E n ij
Controls
Total
MM
n2A
n2U
n2·
Mm
n1A
n1U
n1·
mm
n0A
n0U
n0·
Total
n·A
n·U
n··
2
ij
E n ij
n i n j
n
X2 has distribution with 2 degrees of
freedom under null hypothesis.
2
Cases
• Odds ratio for genotype MM
relative to mm
MM |mm n 2 A n 0 U n 0 A n 2 U
• Affected individual MM |mm times
more likely to have marker genotype
MM than mm.
Cochran-Armitage trend test
•
Assume a multiplicative model of disease
risks:
2
MM |mm Mm |mm
•
The Cochran-Armitage trend test of
association between disease and the
marker SNP is given by
X
2
1
1
p 2 A 2 p 1 A p 2 U 2 p 1U
1
1
n .U
n.A
1
2
n ..
p ij
n ij
n j
X2 has distribution with 1 degree of
freedom under null hypothesis.
2
Controls
Total
MM
n2A
n2U
n2·
Mm
n1A
n1U
n1·
mm
n0A
n0U
n0·
Total
n·A
n·U
n··
2
2
1
1
n .. n 1 . n 2 . n 1 . n 2 .
2
4
where
•
Cases
• Odds ratio for allele M relative to
allele m
M |m
n 1A n 0 U
n 0 . n1 .
n 2 A n 1U 4 n 2 A n 0 U
n1. n 2 . n 0 . n 2 .
1
n 0 A n 1U n 1 A n 2 U 4 n 2 A n 2 U n 0 A n 0 U 2
n 0. n 2.
n 0 . n1 . n1 . n 2 .
2
M |m
• Affected individual
times more
likely to have marker genotype MM
than mm, and M |m times more likely
to have genotype Mm than mm.
Interpretation
A significant result in a test of disease-marker association may imply:
• Marker locus is causative, directly
influencing disease risk: needs to
be established via functional
studies.
• Alleles at marker locus are
correlated with alleles at the
disease locus, but do not directly
influence disease risk: linkage
disequilibrium.
• Population substructure not
accounted for in the analysis,
with different disease and marker
allele frequencies in each
subpopulation.
• False positive signal of
association.
M
Disease
M
D
Disease
M
Pop’n
Disease
Genome-wide significance
• A type I error occurs when we reject the null hypothesis
of no association, when in fact the null hypothesis is true.
• Specify type I error rate – or significance level – at the
design stage of the analysis.
• Lower type I error rate reduces the probability of detecting a
false positive association, but with the penalty of reducing the
power to detect association when it truly exists.
• It is important to correct for multiple testing to maintain
the type I error rate for the experiment overall (i.e. all
the SNPs tested in the association study).
• Genome-wide significance threshold: p<5x10-8.
• Replication is necessary to confirm association.
Logistic regression modelling
• We model the log odds of disease for the ith individual as
pi
log
1 pi
βT x i
where pi is the probability that the ith individual is affected by disease, xi is
a vector of measures of the risk factors, and β is a vector of the
corresponding risk effects.
• Over all individuals, we can obtain estimates of the risk effects by
maximising the log-likelihood
l y x ,β
y
i
i
log p i 1 y i log 1 p i
where yi denotes the phenotype of the ith individual (0 for control and 1
for case).
• Extremely flexible modelling framework: can test for joint effects of risk
factors (multi-locus analysis) and allow for covariates (adjustment for nongenetic effects).
Additive test of association
• Assuming multiplicative model of disease risk (additive on the log scale),
we can model the log-odds of disease for the ith individual by
pi
log
1 pi
0 M x Mi
where βM denotes the log-odds ratio of allele M relative to allele m, and xMi
is an indicator variable that counts the number of M alleles (0, 1 or 2)
carried by the ith individual.
• Test for association by maximising the likelihood of the model, and
comparing with the maximised likelihood under the null hypothesis of no
association (i.e. βM = 0):
X
2
2 l y x M , 0 , M l y x M , 0 , M 0
• X2 has χ2 distribution with one degree of freedom under the null
hypothesis.
• Asymptotically equivalent to Cochran-Armitage trend test.
Genotypic test of association
• To allow for deviations from the multiplicative disease model , we can
model the log-odds of disease for the ith individual by
pi
log
1 pi
0 Mm x Mmi MM x MMi
where βMm and βMM denote the log-odds ratios of genotypes Mm and MM
relative to genotype mm, and xMmi and xMMi are variables indicating that
the ith individual carries genotype Mm and MM, respectively.
• Test for association by maximising the likelihood of the model, and
comparing with the maximised likelihood under the null hypothesis of no
association (i.e. βMm = βMM = 0).
X
2
2 l y x Mm , x MM , 0 , Mm , MM l y x Mm , x MM , 0 , Mm MM 0
• X2 has χ2 distribution with two degrees of freedom under the null
hypothesis.
General disease models
• The same modelling framework can be used to test for association under
alternative disease models, such as heterozygote advantage, recessive and
dominant.
• Testing many different models of association requires correction for
multiple testing.
• We can compare the likelihoods of the genotypic and trend models of
association to test for a deviation from the multiplicative model of disease
risks.
• Trend test is generally most powerful unless there is extreme deviation
from a multiplicative disease model.
• Testing for association at a marker SNP in LD with the causal SNP weakens
the effect of the underlying disease model.
Genotype
M recessive
M dominant
Heterozygote
advantage
MM
1
1
0
Mm
0
1
1
mm
0
0
0
Allowing for covariates
•
•
•
•
•
For complex diseases, we may wish to take account of non-genetic risk factors,
such as exposure to specific environments, or the effects of established genetic
loci.
Assuming multiplicative model of disease risk, we can model the log-odds of
disease for the ith individual by
pi
0 C x Ci M x Mi
log
1
p
i
where βC denotes the effect of a covariate xC, and xCi denotes the value of the
covariate for the ith individual.
Obtain χ2 test of association with one degree of freedom by maximising the
likelihood of the model, and comparing with the maximised likelihood under
the null hypothesis of no association (i.e. βM = 0).
Estimated log-odds ratio of allele M adjusted for the effect of the covariate.
Can be generalised to allow for any number of covariates and general models
of disease risk.
Quantitative traits
• The methodology described here generalises to quantitative
(continuous) traits. It is straightforward to compare the mean
response for each marker genotype by analysis of variance,
assuming a normally distributed trait, within the standard
linear regression framework.
• A powerful strategy is to ascertain individuals from the
extremes of the quantitative trait distribution: cases and
hyper controls.
• We can analyse trait values by linear regression, although this leads to
biased estimates of mean trait values for marker genotypes.
• We can ignore the trait values, and analyse as a standard case-control
sample.
• Are hyper controls representative, or are there polygenic effects
involved?
• This strategy may not be cost effective if phenotyping is expensive
relative to genotyping.
Software
• Contingency table analysis and generalised linear modelling
can be performed using standard statistical software.
• Define indicator variables for specific genetic models from the
observed SNP genotype data.
• Some statistical software packages include specific libraries of
routines to perform genetic analyses (R, STATA)
• Specialised genetic analysis software:
• PLINK. Whole genome association analysis toolset designed to
perform a range of basic, large-scale analyses. Allows for data
management and basic QC analyses. Performs simple case-control
tests of association.
• SNPTEST. Designed for analysis of whole genome association studies.
Allows for flexible single-locus analysis of genotype data allowing for
covariates.
Replication
• To confirm positive association signals from an initial
study, it is essential to replicate the result in
independent samples from the same and/or different
populations.
• Replication of positive association signals has not
proved to be easy: will depend on power of both
initial and replication studies.
• Multi-stage designs: genotype a proportion of
samples with GWAS array and follow-up the
strongest signals of association in the remaining
samples through de novo genotyping.
• Collaboration between international groups studying
the same trait allows for in silico replication.
Meta-analysis
• We can increase power to detect rarer variants of
more modest effect by collecting larger and larger
samples.
• Alternatively, we can combine the results of GWA
studies of the same trait through meta-analysis,
without direct exchange of genotype data.
• Exchange summary statistics for each SNP including
“risk” allele, p-value, odds ratio (effect) and 95%
confidence interval (standard error).
• GWA studies can be combined through meta-analysis
even if genotyped directly for different sets of SNPs
through imputation.
• Software: GWAMA and METAL.
Summary
• Standard statistical procedures available for
the analysis of genotype data from genetic
association studies.
• Logistic regression provides a flexible
framework for modelling SNP association with
disease.
• Signals of association should be validated
through replication and/or meta-analysis.