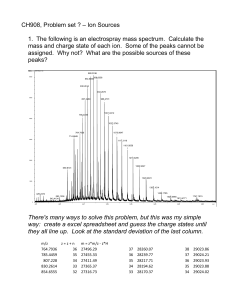
Mass Spectrometry of Proteins and Peptides
advertisement

Mass Spectrometry of Proteins and Peptides Electrospray Ionization(ESI) and Matrix Assisted Laser Desorption Ionization(MALDI) by Matt Fisher Outline • Brief introduction to Proteomics • How ESI and MALDI work • Advantages/setbacks to each method • ESI and MALDI in action Proteomics • “To really understand biological processes, we need to understand how proteins function in and around cells since they are the functioning units.” - Hanno Steen, director of the Proteomics Center at Children's Hospital Boston • 30,000 genes code for 100,000 functional proteins in humans • Extreme cases of a single gene coding for 1,000 proteins! Advances in Protein and Peptide Analysis • Such an enormous task requires every conceivable technique to analyze proteins • The 2002 Nobel Prize for Chemistry was shared between John Fenn, and Koichi Tanaka for their development of ESI and MALDI, respectively Electrospray Ionization Steps to Ionization • Mix liquid sample with polar, volatile solvent • Sample is put through a capillary with a fine tip on the end • A high voltage(~2000 V) is applied to the tip of the capillary, charging the proteins and peptides in the solvent. (Multiply-charged species common in ESI) • Mixture is pushed through to an evaporation chamber Electrospray Ionization Electrospray Ionization Evaporation Chamber • The mixture starts out in “large” droplets. The addition of nitrogen gas and heat begins to evaporate the solvent in the droplets • The droplets get smaller and the charged molecules get closer together and repel, splitting into smaller droplets(Coulombic fission) • The process continues until each droplet consists of a single molecule that is charged • Molecules then enter a mass analyzer such as a time of flight(TOF) tube to measure m/z Matrix Assisted Laser Desorption Ionization (MALDI) •Liquid sample first mixed with an excess of matrix on a MALDI plate • The liquid in the mixture evaporates in open air, with some of the sample incorporated into fine crystals of the matrix • The matrix is a UV-absorbing species, usually of low molecular weight MALDI • MALDI plate is put into a high vacuum chamber and the laser is fired in bursts at crystals on the spots on the plate • At the right wavelength the crystals are irradiated and sublime. Energy is transferred to the analytes which are now in the gas phase • These protonated ions are accelerated into a mass analyzer such as a TOF tube Advantages/Disadvantages to ESI Advantages Disadvantages • High accuracy • Large mass range • Can be coupled with liquid chromatography to separate samples further • Fast • Auto run with sampler or direct injection • Soft ionization • Complicated spectra • salts drown signal and take time to remove from the machine • A high intensity peak can eclipse smaller intensity peaks • Fine tuning work: flow rate, solvent/sample ratios, etc to get the analytes to ionize Advantages/Disadvantages to MALDI Advantages Disadvantages • Preferable for large molecules • Quick, quick, quick! • Sensitive to small amounts of sample • Easy spectra • Accurate • Not affected by salts • Soft ionization • Fine tuning: spotting plate, getting good crystals, adjusting intensity of laser, finding crystals on plate with sample • Low shot to shot reproducibility • Short sample life ESI Spectra CPV Bromelin_30min Max. Entropy Deconvolution Intens. x10 5 3 2 1 0 0 5 10 15 20 25 Time [min] CPV_B_30min_41_01_2213.d: TIC +All MS Intens. +MS, 12.0-12.9min #(1013-1091) 300 831.8 250 624.1 864.9 200 979.3 943.5 150 100 50 0 200 400 600 800 1000 1200 Intens. x10 4 1.0 0.8 1400 1600 m/z +MS, 12.0-12.9min #(1013-1091), Deconvoluted (maximum entropy) Intact protein(VP2): 64,567.2 Da 64567.2 Digested protein(VP3): 62,315.8 Da 62615.8 0.6 0.4 65664.7 0.2 67369.8 51835.7 53151.8 57351.0 55110.0 69381.7 60727.7 0.0 50000 52000 54000 56000 58000 60000 62000 64000 66000 68000 m/z Intens. x10 5 ESI Spectra CPV Bromelin_150 min Max Entropy Deconvolution 3 2 1 0 0 5 10 15 20 25 Time [min] CPV_B_120min_42_01_2217.d: TIC +All MS Intens. +MS, 12.0-12.6min #(1019-1073) 831.8 300 624.1 864.9 979.3 200 100 0 200 400 600 800 1000 1200 Intens. x10 4 1400 1600 m/z +MS, 12.0-12.6min #(1019-1073), Deconvoluted (maximum entropy) 1.25 62614.4 Intact protein(VP2): 64,567.6 Da 64567.6 1.00 Digested protein(VP3): 62,614.4 Da 0.75 0.50 65666.0 0.25 61510.5 67372.1 51836.7 53151.7 57344.5 54833.0 69805.9 59841.6 0.00 50000 52000 54000 56000 58000 60000 62000 64000 66000 68000 m/z ESI Spectra CPV Bromelin_23 hr Max. Entropy Deconvolution Intens. x10 5 2.5 2.0 1.5 1.0 0.5 0.0 0 5 10 15 20 25 Time [min] CPV_B_23hrs_43_01_2221.d: TIC +All MS Intens. +MS, 12.0-12.6min #(1024-1070) 831.8 300 624.1 864.8 979.3 200 1099.6 100 0 200 400 600 800 1000 1200 Intens. x10 4 1400 1600 m/z +MS, 12.0-12.6min #(1024-1070), Deconvoluted (maximum entropy) 1.25 Intact Protein(VP2): 64,568.8 Da 1.00 Digested Protein(VP3): 62,615.8 Da 62615.8 64568.8 0.75 0.50 0.25 65663.8 61504.5 67375.2 51835.0 53148.6 57353.8 54840.0 59839.4 69389.9 0.00 50000 52000 54000 56000 58000 60000 62000 64000 66000 68000 m/z 2007_05_31 MALDI analysis of CPV reacted with Trypsin @ 45°C 5 min Sources • C.Nelson, E.Minkkinen, M. Bergkvist, K.Hoelzer, M. Fisher, B. Bothner, and C.Parrish (2008). “Detecting Small Changes and Additional Peptides in the Canine Parvovirus Capsid Structure”. J. Virol. 82: 10397-10407 • http://www.magnet.fsu.edu/education/tutorials/tools/ionization_esi.html • H. Steen, M. Mann (2004). “The Abc’s (and xyz’s) of Peptide Sequencing”. Nature Reviews Molecular Cell Biology 5, 699-711 • http://www.childrenshospital.org/cfapps/research/data_admin/Site602/mainpageS602 P0.html • F.Witzmann, J. Li (2002). “Proteomics: Core Technologies and Applications in Physiology”. American Journal of Physiology – Gastrointestinal and Liver Physiology. 10.1152 • http://www.innovadyne.com/Assets%20Doc/MALDI%20spots%20Biomek%20plate.jpg