Exam-in-protein-chemistry-NKED15_TFKE46_20130315
advertisement

Exam in protein chemistry NKED15/TFKE46 2013-03-15
NOTE question 4 is worth 23 points!!
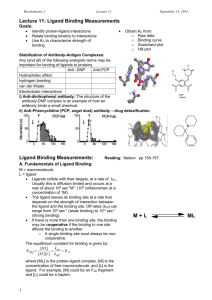
1 a) Keratin is based on a seven residue or heptad repeat. Describe the properties of this
sequence that favour coiled-coil structures.
(2p)
b) Several connective tissue diseases have been identified as arising from mutations in genes
encoding collagen chains. Most common are single base mutations that result in the
substitution of glycine by a different residue. Describe the molecular basis of these diseases
using your knowledge of structure and stabilizing interactions in collagen.
(3p)
c) Membrane proteins are often divided in two groups depending on their position in the
membrane. What are the names for these two groups?
(2p)
d) Only a few percent of all protein structure coordinates deposited in the Protein Data Bank
comes from membrane proteins. What is the reason for the low percentage of determined
structures of membrane proteins?
(2p)
d) Hydropathy plots of two different membrane proteins of similar size were performed
(Figure 1). Which hydropathy profile belongs to which protein? Give an explanation to the
different hydropathy profiles.
(4p)
1
A
B
C
D
Figure 1A-D Three-dimensional structure of two membrane proteins and it corresponding hydropathy plot.
2. a) Enzyme catalysis is often very efficient which can be exemplified by ester hydrolysis.
The enzyme catalyzed reaction had a kcat = 1.1 x 103 s-1. The corresponding uncatalyzed
reaction had a rate constant of 1.2 x 10-3 M-1s-1. Calculate the catalytic effect and give
comments to the result.
(3p)
b) Draw an energy profile for an enzyme catalyzed reaction and illustrate what difference in
energy levels that can be calculated from kcat/Km.
(3p)
c) For an enzyme it was suspected that an active-site Tyr was forming a critical H-bond with
substrate in the transition state of the enzymatic reaction. By site-directed mutagenesis this
Tyr was replaced by a Phe and then kcat/Km-values were measured for the wild type and
mutant. The values turned out to be 88 M-1s-1 for the wild type and 0.1 M-1s-1 for the mutant.
Do these data give support to the assumed H-bond stabilization of the transition state?
Support your conclusions with relevant calculations.
(kobs = kBT/h exp(-ΔG#/RT); where kobs is a rate constant, R=1.99 cal/moldegree, h=1.58x1034
cals, kB=3.3x10-24cal/degree, T=298 K).
(4p)
2
3 a) Discuss why proteins are only marginally thermodynamically stable (5 – 15 kcal/mol).
Moreover, how comes that the contribution to the protein stability from a H-bond is only
approx. 1 kcal/mol, when the corresponding bond between two water molecules is 5 kcal/mol?
(2p)
b) How can a chaperone like GroEL prevent a folding molecule from aggregation?
In what state is the folding protein most prone to aggregation and why is that?
(2p)
c)The folding mechanism of the SH3 domain has been studied by φ-value analysis. Describe
how ΔΔG# can be determined for the transistion state between I and F (TS(IF) in Figure
2).(NOTE: Figure 2 (without figure text) is also available in colour in Appendix 1)
(2p)
3
Figure 2. Changes in ΔG used to calculate the φ-value, along with changes in free energy upon point mutation,
ΔΔG, along the folding pathway.Values of G are referenced with respect to the unfolded state U that is
arbitrarily assigned a value of 0. TS(UI) and TS(IF) denote the rate-limiting transition states between states U,I
and I,F respectively. Inset for the A39V/T47S/N53P/V55L mutant shows the pair of DG profiles ('pseudo-wt' and
mutant) from which ΔΔG values are obtained, with the free energies of U states both assigned arbitrarily to 0.
I is a folding intermediate and the top figure shows just in a general way how the comparisons are made for a
general state denoted A. The mutations A39V/N53P/V55L do not affect the protein at all compared to the wildtype and the protein variant with these mutations are regarded as a pseudowild-type form. Thus, the mutations to
consider (shown in bold face) here are from top to botton in the figures above: T47S, R40T, L3A, E5V and F20L.
4
d) What can be concluded from the φ-values about the formation of the β-turn between βstrands 3 and 4 (as probed by positions 40 and 47 (Figure 3)) as well as the folding of β-strand
1 (as probed by positions 3 and 5 (Figure 3))?
(2p)
e) From NMR-data there is evidence that the intermediate I has a non-native
hydrophobic core. What does the F20L mutant tell us regarding that in this study?
(2p)
Figure 3. Schematic representation of the secondary structure of a homology model of theA39V/N53P/V55L
Gallus gallus Fyn SH3 domain (the 'pseudo-wild-type' in this study) in the native state F, featuring the
characteristic SH3 domain β sandwich
fold formed by the terminal (strands β1, β5) and the approximately orthogonal central β-sheets (strands β2,
β3, β4), along with α 310-helical turn.
Residues mutated for φ-value analysis are shown in ball-and-stick representation.
5
4 ) Methyltransferases is a large protein family with different functions in the cell, e.g
detoxification of substances. A protein structure of one of the member of this family is
illustrated below showing the three-dimensional structure, sequence and topology diagram
(Figure 4 A, B, and C)
A
B
6
C
Figure 4 A) Three-dimensional structure of a methyltransferase, B) topology diagram of the same
methyltransferase and C) secondary structure and amino acid of the same methyltransferase.
a) The above figure (Figure 4A) shows the structure of a well-known protein motif. What
is the protein motif called?
(2p)
b) To function this protein bind a cofactor called S-adenosylmethionine illustrated below
(figure 5). Suggest a binding site for the cofactor using the topology diagram
illustrated above (figure 4B)
(2p)
Figure 5 Structure of S-adenosylmethionine
c) Two emission spectra of the methyltransferase using an excitation wavelength of 295
nm under native conditions (0 M GuHCl) and denatured condition (6M GuHCl) were
performed (figure 6). What conclusion can be drawn regarding the positions of the Trp
7
in the methyltransferase? Trp fluorescence is often used to monitor stability of a
protein. Why is this method not suitable in this case?
(4p)
Fluorescence intensity A.U
14000
12000
10000
8000
6000
4000
2000
0
320
340
360
380
400
Wavelength (nm)
Figure 6. Emission spectra of methyltransferase under native ( filled circle) and denatured (open cicle)
conditions.
d) Instead of using Trp fluorescence, an alternative method was used. The extrinsic
fluorescent probe, ANS (figure 7) was added to the samples and the thermal stability
was monitored (Table 1). Calculate the stability of the protein and interpret the data in
terms of structural changes.
(3p)
Figure 7 Extrinsic fluorescent probe, Anilino-naphtalene sulphonate, ANS
8
Table 1 Experimental data monitoring the fluorescence intensity at 475 nm at various temperatures
Temperature
18
20
22
24
26
28
30
32
34
36
38
40
42
44
46
48
50
52
54
56
58
60
62
64
66
68
70
72
74
76
78
80
Fluorescence Intensity at 475 nm
27560
27620
26350
26690
26030
26140
26280
25700
24690
23820
22740
22080
20240
19070
16430
13470
9440
7570
4740
3490
3520
3430
3170
2180
2410
1870
1190
1070
500
140
190
180
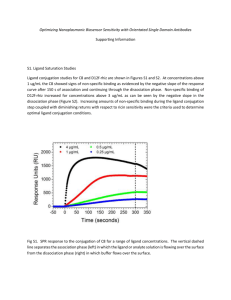
e) To calculate the binding of the cofactor to the protein a ligand binding assay was
performed using equilibrium dialysis. Describe the principle of two other methods you
can use to measure ligand binding.
9
(4p)
f) To analyse the binding of a ligand to the protein an equilibrium dialysis experiment
was performed. The protein concentration was kept constant at 4 mg/ml and the
molecular weight of the protein was 40000 Da. The experimental data for the ligand
binding assay is illustrated below (Table 2). Calculate the dissociation constant and
number of binding sites. Give an explanation of what type of binding it is.
(4p)
Table 2 Experimental data from equilibrium dialysis
Total ligand concentration (mM)
0.010
0.020
0.050
0.075
0.100
0.150
0.200
0.400
0.700
1.000
Bound ligand concentration (mM)
0.005
0.009
0.021
0.030
0.039
0.047
0.058
0.067
0.075
0.099
g) The mutation Y240C interacts with a residue in the vicinity and cause stabilization of
the protein. Give a reasonable cause for this stabilization and identify the interacting
residue.
(4)
10